Introduction
P. aeruginosa is a non-fermentative Gram-negative bacteria widely distributed in nature and can survive on a wide variety of surfaces and in hospital environment, as the wards encourage bacterial growth (Arora et al., 2011). P. aeruginosa is responsible for about 10% -20% of nosocomial infections as bacteraemia and sepsis in ICU, cystic fibrosis, pneumonia, urinary tract infections, burn infection and wound infection (Carmeli et al., 1999). Multidrug-resistant (MDR) P. aeruginosa phenotype is defined as resistant to one anti-microbial agent in three or more anti-pseudomonal anti-microbial classes (carbapenems, fluoroquinolones, penicillins /cephalosporins and aminoglycosides) (Magiorakos et al., 2011 and Maria et al., 2011). MDR P. aeruginosa are particularly problematic because the organism is inherently resistant to many drug classes and is able to acquire resistance to all effective antimicrobial drugs (Gad et al., 2007). MDR P. aeruginosadevelops resistance by various mechanisms like multi-drug resistance efflux pumps, biofilm formation, production of β-lactamases and aminoglycoside modifying enzymes. The risk for acquiring MDR organisms may be related to the number of carriers in the same ward as well as to individual risk factors, such as patient characteristics and in-hospital events (invasive devices and antibiotic treatment) (Carmeli et al., 2002). Extended spectrum beta lactamases (ESβLs) have been described in P. aeruginosa only recently. β-lactamases described in P. aeruginosa belong to various families as TEM and SHV types which are common among Enterobacteriaceae, PER type, VEB type which have been reported from various parts of the world (Amutha et al., 2009). Typing techniques are essential for epidemiological purposes to establish a clonal relationship between different strains isolated from patients and those isolated from surrounding environment and medical team. Phenotypic methods such as biotyping, antibiotyping, pyocin typing and serotyping and molecular methods such as plasmid profile analysis and PCR were used for this purpose. This study was planned to delineate prevalence and resistance patterns of P. aeruginosa and MDR phenotype in nosocomial infection patients, and to screen for ESβLs producing P. aeruginosa in Menofia University Hospitals (MUH). Also to characterize clinical and environmental isolates of P. aeruginosa by plasmid profile analysis, antibioyping and pyocin typing, aiming for a better management, reducing both mortality and costs.
Materials and Methods
This study was conducted during the period from May 2009 to July 2011 and included 287 inpatients (group I) (165 males and 122 females) admitted to different wards and units of MUH and having different nosocomial infections, their ages ranged from 0 to 90 years. A total of 287clinical specimens were examined; 78 urine specimens, 46 ear swabs, 25 wound swabs and 65 burn swabs. 51 hand, throat and nasal swabs were collected and processed from 17 medical staff (3 swabs each) (group II) working at MUH. 40 environmental samples were taken from surgical instruments, dressings, bath, suction devices, floors, walls, beds, commodes, sinks and antiseptic solutions. Samples were inoculated onto blood agar, nutrient agar, CLED agar and MacConkey agar (Oxoid, England). Colonies grown on different media were subjected to further morphological and biochemical identification. Suspected P. aeruginosa colonies were identified according to standard microbiological methods (Koneman et al., 2006). Antibiotic sensitivity and antibiotyping of P. aeruginosa clinical and environmental isolates were determined by disc diffusion method of Kirby Bauer on Muller-Hinton agar (Oxoid, England) and interpreted according to the method described by the Clinical and Laboratory Standard Institute (CLSI, 2009). In our work MDR P. aeruginosa was detected as resistant to one anti-microbial agent in three or more anti-pseudomonal anti-microbial classes (Magiorakos et al., 2011 and Maria et al., 2011). Strains having the same resistance patterns (antibiotype) considered to be from the same clone. Phenotypic confirmatory tests recommended by the CLSI for ESβLs detection were carried to assess the prevalence of ESBL. The double — disc synergy test was done, organisms were swabbed onto a Mueller — Hinton agar plate (Oxoid, England). Disc containing amoxicillin — clavulanate (20 µg â„ 10µg respectively) was placed in the center of the plate. Discs containing one of the oxyimino —β— lactam antibiotics (cefotaxime, ceftazidime, ceftriaxone and aztreonam) were placed 30 mm (center to center) from amoxicillin — clavulanate disc and incubated at 35oC for 24 hours. A clear extension of the edge of the oxyimino β-lactam inhibition zone towards the disc containing clavulanate was interpreted as synergy indicating a positive result. Another phenotypic confirmatory test, cephalosporin/clavulanate combination discs this requires use of both ceftazidime – clavulanic acid (30 µg ∕10 µg) and cefotaxime -clavulanic acid (30 µg ∕10µg) discs. The discs were applied to the plates using a sterile forceps and incubated at 35°C for 18 hours. An increase in the zone diameter of ≥5 mm in the presence of CA, compared to ceftazidime or cefotaxime alone, is taken to be a phenotypic confirmation of ESβL production (CLSI, 2009). Plasmid DNA analysis for all P. aeruginosa isolates were done. Plasmids were extracted as previously described by alkaline lysis method (Sambrok et al., 2001 and Ranjbar et al., 2007). Extracted plasmids were then separated by electrophoresis, stained with ethidium bromide and visualized under UV illumination. The strains were grouped depending on the size and number of the plasmid DNA bands. Pyocin typing was done by Gillies and Govan technique (Fyfe et al., 1984). Each isolated strain was streaked diametrically across the surface of tryptone soya agar to give an inoculum width of approximately 1 cm. The growth was exposed to chloroform for 15 minute. The plate was then opened exposing the medium to air for few minutes. Five standard indicator strains were grown in nutrient broth were then streaked on to the medium at right angles to the line of the original inoculum. The plate was then incubated at 37°C for 18hrs. Any pyocin produced by the original tested strains diffused into the medium during the first period of incubation and then exerted their inhibitory action on the indicator strains during the second incubation. Plates were observed for growth inhibition of indicator strains.
Results
The present study included 287 inpatients, out of them 283 clinical isolates were recovered. P. eruginosa accounted for 19 % (54 of 283) of nosocomial infections isolates. Only 3 P. aeruginosa strains were isolated from environmental sites. In our work, P. aeruginosa wasn’t detected from medical staff swabs. P. aeruginosa infections were common in males (66.7 %) than female (33.3%) cases and their mean age was (44.6±25.8). In our study, P. aeruginosa mostly isolated from burn unit (32.3%), ICU (16.7%) then urology department (18.75%) (Table I) and (Figure I).
Table (I): Distribution of P. aeruginosa Isolates among Different Departments
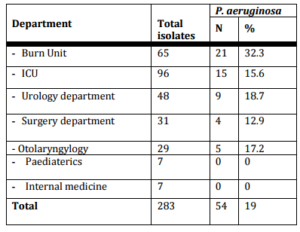
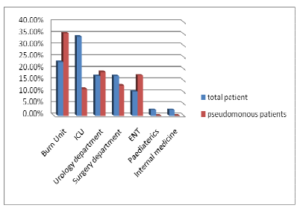
Figure (I): Distribution of P. aeruginosa Isolates among Different Departments
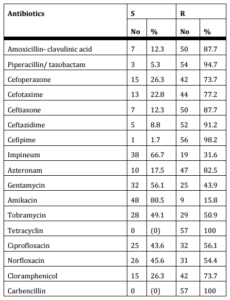
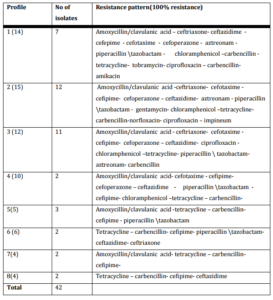
Table (II) summarizes the resistance patterns of 57 P. aeruginosa strains (54 clinical and 3 environmental isolates). Amikacin was the most effec¬tive drug against all P. aeruginosa isolates showed maximum sensitivity (80.5%) followed by imipenem (66.7%) and gentamicin (56.1%). On the other hand, P. aeruginosa had high resistance rates to cefepime (98%) followed by piperacillin tazobactam (94.7%), ceftazidime (91%) and ceftriaxon and amoxicillin- clavulnic acid (87% each). All isolates were totally resistant to carbincillin and tetracycline.
Table (II): Antibiotic Susceptibility Tests of P. aeruginosa Isolates, 54 Clinical Isolates from Group I and 3 Environmental Isolates by Disc Diffusion Method
Antibiogram categorized 57 P. aeruginosa isolates into 23 different antibiotypes, (1-8) antibiotypes including 42 strains are the most prevalent (table III). 3 environmental isolates and 9 clinical isolates were included in antibiotype (2).
Table (III): Groups of P. aeruginosa Isolates that Have the Same Antibiotype
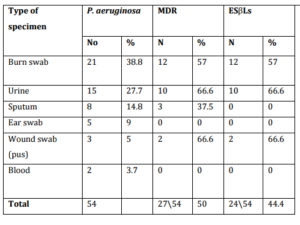
In our work, 52% of P. aeruginosa were MDR and were categorized in antibiotype 1, 2 and 3. Those 30 MDR P. aeruginosa isolates were distributed as follow 27 were clinical isolates and accounted for 9.5 % (27283) of all nosocomial infections and 3 were environmental isolates. In our work 45.6% P. aeruginosa (24 clinical isolates and two environmental) were positive for ESβLs production, and all were MDR (26 of 30) 86.6%. In this study, out of total 54 P. aeruginosa isolates, 2154 (38.8%) were from burn wound, 15 (27.7%) from urine, 8 (14.8%) from sputum, 5 isolates from ear discharge, 3 from surgical wound and 2 isolates from blood. MDR – ESβLs producing P. aeruginosa (24) isolates were isolated from burn wound 12 of 21 (57%), 10 of 15 (66.6%) from urine and 2 of 3 (66.6%) from surgical wound (Table IV).
Table (IV): Distribution of P. aeruginosa, MDR and ESBLs Producing Isolates in Clinical Specimens
On analysis of plasmid profile, out of 57 P. aeruginosa 27 strains (47.4%) harbored plasmids and were categorized into 7 different plasmid profiles (A-G). The sizes of plasmids varied from 1.6 to 140 kb (Figure 3). Two environmental and three clinical isolates had plasmid profile (A). While studying the correlation between plasmid profile and ESβLs production of P. aeruginosa, it was found that 16 of 26 (61.5%) ESβLs producer strains had plasmids and were categorized in 4 plasmid profiles (A,B,C and D) (Table V).
Table (V): Relation between Resistance Patterns and Plasmid Profiles among 16 of 26 (61.5%) MDR-ESβLs Producer P. aeruginosa
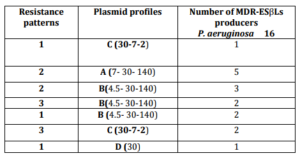
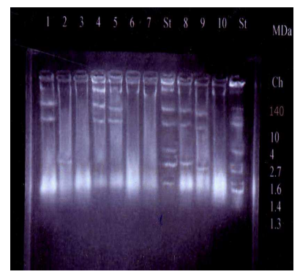
Figure (II): Plasmid Profile of Some of the Studied P. aeruginosa Isolates
From left to right: Lane 1, 4, 5 shows 3 plasmids (140-30-7 MDa). Lane 2, 6 shows 1 plasmid (2.2MDa). Lane 9, 10 shows 3 plasmids (30-7-2 MDa). Lane 7, 11 shows no plasmids. Lanes 8 & 12 (standard strain) show 7 plasmids (140-10-4.8-2.7-1.6-1.4-1.3MDa).
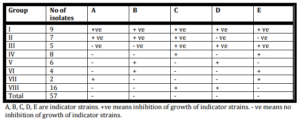
In the current study P. aeruginosa isolates (57) were typed by pyocine typing method. The results of pyocin typing classified P. aeruginosa isolates into 8 groups (I-VIII) (Table VI). Pyocine typing method couldn’t discriminate MDR or ESBLs P. aeruginosa isolates in unique groups.
Table (VI): Pyocin Typing of P. aeruginosa Isolates
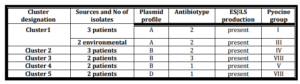
Five clusters could be demonstrated among our MDR P. aeruginosa isolates. Each cluster had the same antibiotic sensitivity pattern and plasmid profile (Table VII), while cluster one isolates had different pyocin type. Two environmental isolates were identical by antibiotyping and plasmid profile to 3 clinical isolates and present in cluster 1 (Table VII).
Table (VII): Clusters of MDR P. aeruginosa Isolates that Have the Same Antibiotic Sensitivity Pattern, Plasmid Profile and β-Lactamase Production and Their Relations to Pyocin Groups
Discussion
P. aeruginosa is a well-recognized nosocomial pathogen that can cause severe infections in hospitalized patients. A total of 57 P. aeruginosa strains were isolated (54 clinical and 3 environment isolates). P. aeruginosa and its MDR phenoype accounted for 19 % and 9.5% respectively of nosocomial infections in MUH. This is nearer to 18% rate of nosocomial infection reported by Gad et al., (2007) from three hospitals in Minia, Egypt. Lower incidence reported by Khan et al.,(2008) in Pakistan 6.67%. In our wok P. aeruginosa and MDR- ESβLs producer P. aeruginosa infections were most common in burn wound infection followed by urinary tract, respiratory tract and wound infections. The highest incidence ofP. aeruginosa was found in burn unit and ICU. In concordance with our result Aly, (2005) who founded that P.aeruginosaaccounted for (44.4%) of infections in burn unit at MUH. Also Pathmanathan et al., (2009) concluded that P. aeruginosais a nosocomial pathogen often isolated from burn infections. ICUs are generally considered epicenters of (MDR) organisms (Ramprasad et al., 2010). MDR P. aeruginosa had emerged in Egypt in recent years and seen mainly in nosocomial infections (Ali et al., 2009). In this study the isolated P. aeruginosa were investigated for MDR and ESβLs production. We found high prevalence of MDR P. aeruginosa (52%) and ESβLs producer strains (45.6%) were found in MUH. This high rate of MDR, has been reported elsewhere in previous studies. In Turkey, Unan et al., (2000) reported that 60% of his P. aeruginosa isolates were MDR. In Egypt, Gad et al., (2007) observed high levels of MDR P. aeruginosa and that β-lactamase production is the main mechanism of resistance (36% were MDR and 95% were ESβLs producer) On the other hand, Zahra (2011) in Iran detected lower levels as 30% of their isolates were MDR and only 9.2% were ESβLs producer. Egypt is among the countries that reported high rates of antimicrobial resistance (El-Kholy et al., 2003). MDR P. aeruginosa develops resistance by various mechanisms like multi-drug resistance efflux pumps, production of β-lactamases, aminoglycoside modifying enzymes, and decrease outer membrane permeability (Maria et al. 2011). Amikacin and imipenem were the most effec-tive drugs against P. aeruginosa. The effectiveness of amikacin (80.5% sensitive) over imipenem (66.7% sensitive) against P. aeruginosa was corroborated by data from recent researches as Gad et al., (2007). Earlier studies reported that imipenem was the most effective antibiotic against P. aeruginosa Gales et al., (2002). However, recent studies demonstrated the evolution of imipenem -resistant strains of P. aeruginosa (Maria et al., 2011). Carbapenems were considered to be the treatment of choice against serious ESβLs associated infections, however resistance to carbapenems, especially in P. aeruginosa, results from reduced levels of drug accumulation, increased expression of pump efflux or production of β-lactamases (Maria et al., 2011). Our study revealed moderate activity of ciprofloxacin 43.6%. There were variable data about ciprofloxacin action .Similar results about cipro against P. aeruginosa were detected by Gad et al., (2007) , on the other hand Zahra (2011) reported that ciprofloxacin exhibited high susceptibility pattern (73.6%). In our research, high resistance rates exhibited by P. aeruginosa against cefepime (98%), ceftazidime (91%) and ceftriaxon (87%). This may be due to high ESβLs production in our P. aeruginosa isolates used β -lactamase-mediated resistance against cephalosporins. In contrast, cephalosporins tested in a study conducted in Nigeria, showed that 90% of the isolates were sensitive to it (Oni et al., 2002). This discrepancy can be attributed to the continuous development of MDR strains in different parts of the world, also a considerable geographic difference in the prevalence of ESβLs in different countries. In general, in our work high resistance patterns were exhibited by most of P. aeruginosa isolates. Typing of nosocomial isolates is essential for determining the epidemiology of nosocomial infections and aiding in the design of rational pathogen control methods. Antibiotyping was very discriminative in our work, 23 different resistance patterns were identified among our P. aeruginosaisolates. Profiles (1-8) were prevalent and included 42 strains. MDR were categorized in antibiotype 1, 2 and 3. Antibiogram is a sensitive phenotypic marker, however it has the disadvantage of being non reproducible in many instances due to the exchange of R factor among isolates (Ramprasad et al., 2010). In our work, 27 (47.4%) isolates had plasmid with 7 different plasmid profiles (A-G). This coincides with that reported by Nikbin et al., (2007) in Iran detected plasmids in 39.8% of his P. aeruginosa isolates. In our study, there were a correlation between plasmid profile and ESβLs production of P. aerugionsa isolates, it was found that 16 (61.5%) of the 26 ESβLs isolates harbor plasmids of different sizes. Nearer results reported by Ali et al., (2009) as 55% of his isolates were ESβLs producers. Lower rates were obtained by Anjum and Mir, (2009) in Pakistan who found that out of 100 isolates, 33 (33%) were found to be ESβLs producers. In our work, 10 strains of the ESβLs producers were plasmidless and β -lactamase production in these strains could be chromosomally mediated. A finding which was explained by Bradford, (2001). In our study, pyocin typing didn’t differentiate P. aeruginosa isolates belonging to different sources. There were 8 pyocin groups in our work but there were no difference between clinical and environmental isolates. Contrary to our result, Ramprasad et al., (2010) found pyocine typing is a very sensitive method of typing. A possible common source of infection in our study may be hospital environment, as two environmental isolates were identical by two typing method to 3 clinical isolates. Environmental sources may have a significant role in the transmission of P. aeruginosa (Bradford, 2001). This study highlighted the need for additional attention to the disinfection of inanimate objects in the hospital environment to limit the transfer of P. aeruginosa. A close relationship was found between the presence of certain plasmids and characteristic patterns of antibiotic resistance as five clusters could be demonstrated among our P. aeruginosa isolates, each had the same antibiotic sensitivity pattern, plasmid profile and β-lactamase production. These results are nearer to Bradford, (2001)who reported that combination of multiple typing methods may lead to more precise results and more efficient tracing of the source of infection. In conclusions, our results clarified that threats of MDR and ESBLs P. aeruginosa become of major concern in our hospital and implementation of infection control strategies are major concerns to avoid the spread of this threat. Environmental sources may have a significant role in spread of MDR P. aeruginosa among hospitalized patients. Combination of simple typing methods as antibiotyping with plasmid profiling may be the cheapest and effective method for tracing source of infection.
References
Al-Jasser, A. M. (2006). ‘Extended-Spectrum Beta-Lactamases (ESβLs): A Global Problem,’ Kuwait Medical Journal, 38 (3) 171-185.
Ali, A. M., Abbasi, S. A. & Ahmed, M. (2009). ‘Frequency of ESβLs Producing Nosocomial Isolates in a Tertiary Care Hospital in Rawalpindi,’ Pakistan Armed Forces Medical Journal, 12(3) 23-28.
Aly, A. A. (2005). ‘Hospital-Acquired Infection in Burn Unit in Menoufiya University Hospital,’
Amutha, R. P., Murugan, T. & Renuga devi, M. P. (2009). “Studies on MDR P. aeruginosa from Pediatric Population with Special Reference to Extended Spectrum Beta Lactamase,” Indian Journal of Science and Technology, (2) 6846 -6851
Publisher
Anjum, F. & Mir, A. (2010). “Susceptibility Pattern of P. aeruginosa against Various Antibiotics,” African Journal of Microbiology Research, 4 (10) 1005-1012
Publisher – Google Scholar
Arora, D., Jindal, N., Kumar, R. & Romit, M. (2011). “Emerging Antibiotic Resistance in Pseudomonasa Challenge,ʼInternational Journal of Pharmacy and Pharmaceutical Sciences, 3( 2) 1488-1491
Publisher
Bradford, P. A. (2001). “Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of this Important Resistance Threat,” Clinical Microbiology Reviews, 14(4) 933—951.
Publisher – Google Scholar – British Library Direct
Carmeli, Y. N., Eliopoulos, G. M. & Samore, M. H. (2002). “Antecedent Treatment with Different Antibiotic Agents,”Emerg. Infect. Dis., (8) 802—807.
Publisher – Google Scholar – British Library Direct
Carmeli, Y. N., Troillet, G., Eliopoulos, G. M. & Samore, M. H. (1999). “Emergence of Antibiotic-Resistant Pseudomonas Aeruginosa: Comparison of Risks Associated with Different Antipseudomonal Agents,” Antimicrob. Agents Chemother, (3) 1379—1382
Publisher – Google Scholar – British Library Direct
CLSI (2009). ‘Performance Standards for Antimicrobial Susceptibility Testing,’ 18th informational supplement.
Corona-Nakamura, A. L., Miranda-Novales, M. G., Leanos-Miranda, B., Portilo-Gomez, L., Hernadez-Chavez, A., Anthor-Rendon, J. & Aguilar-Benavides, S. (2001). “Epidemiologic Study of Pseudomonas Aeruginosa in Critical Patients and Reservoirs,” Archives of Medical Research, (32) 238-242.
Publisher – Google Scholar – British Library Direct
El-Kholy, A., Baseem, H., Hall, G. S., Procop, G. W. & Longwoth, D. L. (2003). “Antimicrobial Resistance in Cairo, Egypt, 1999-2000: Survey of 5 Hospitals,” Journal of Antimicrobial Chemotherapy, (51) 625-630.
Publisher – Google Scholar – British Library Direct
Fyfe, J. A., Harris, G. & Govan, J. R. (1984). “Revised Pyocin Typing Method for Pseudomonas Aeruginosa,” Journal of Clinical Microbiology, (20) 47—50.
Publisher – Google Scholar
Gad, G. F., El- Domany, R. A., Zaki, S. & Ashour, H. M. (2007). “Characterization of Pseudomonas Aeruginosa Isolated from Clinical and Environmental Samples in Minia, Egypt: Prevalence, Antibiogram and Resistance Mechanisms,” Journal of Antimicrobial Chemotherapy, (60) 1010—1017
Publisher – Google Scholar – British Library Direct
Gales, A. C., Sader, H. S. & Jones, R. N. (2002). “Urinary Tract Infection Trends in Latin American Hospitals: Report from the SENTRY Antimicrobial Surveillance Program (1997—2000),” Diagn Microbiol Infect Dis, (44) 289—99
Publisher – Google Scholar
Gill, M. M., Usman, J., Kaleem, F., Hassan, A., Khalid, A., Anjum, R. & Fahim, Q. (2011). “Frequency and Antibiogram of Multi-drug Resistant Pseudomonas Aeruginosa,” Journal of the College of Physicians and Surgeons Pakistan, 21 (9) 531-534
Publisher – Google Scholar
Inan, D., Ogunc, D., Gunseren, F., Colak, D., Mamikoglu, L. & Gultekin, M. (2000). ‘The Resistance of Pseudomonas aeruginosa Strains Isolated from Nosocomial Infections against Various Antibiotics,’ Mikrobiyol Bult (34) 255-60
Google Scholar – British Library Direct
Khan, J. A., Iqbal, Z., Rahman, S. U., Farzana, K. & khan, A. (2008). “Prevalence and Resistance Pattern of Pseudomonas Aeruginosa against Various Antibiotics,” Pak. J. Pharm. Sci., 21(3) 311-315.
Publisher – Google Scholar
Koneman, E., Winn, W. J., Allen, S., Janda, W., Procop, G., Woods, G. & Schreckenberger, P. (2006). ‘Koneman’s Color Atlas and Textbook of Diagnostic Microbiology,’ Sixth edition, chapter 7 p264-269. Lippincott Williams & Wilkins, London.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Grundmann, H., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Suetens, C., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T. & Monnet, D. L. (2011). “Multidrug-Resistant (MDR), Extensively Drug Resistant (XDR) and Pandrug-1 Resistant (PDR) Bacteria in Healthcare Settings. Expert Proposal for a Standardized International Terminology,” Available online at www.escmid.org.
Publisher – Google Scholar
Moniri, R. & Tavajjohi, Z. (2011). “Detection of ESBLs and MDR in Pseudomonas Aeruginosa in a Tertiary-Care Teaching Hospital,” Iranian Journal of Clinical Infectious Diseases, 6(1) 18-23
Publisher – Google Scholar
Nikbin, V. S., Abdi-Ali, A., Feizabadi, M. M. & Gharavi, S. (2007). “Pulsed Field Gel Electrophoresis & Plasmid Profile of Pseudomonas Aeruginosa at Two Hospitals in Tehran, Iran,” Indian J Med Res , (126) 146-151.
Publisher – Google Scholar – British Library Direct
Oni, A. A., Nwaorgu, O. G. B., Bakare, R. A., Ogunkunle, M. O. & Toki, R. A. (2002). ‘The Discharging Ears in Adults in Ibadan, Nigeria; Causative Agents and Antimicrobial Sensitivity Pattern,’ Afr J Clin Exp Microbiol , (3) 3-5.
Pal, R. B., Rodrigues, M. & Datta, S. (2010). “Role of Pseudomonas in Nosocomial Infections and Biological Characterization of Local Strains,” j Biosci Tech, l 1 (4) 170-179
Publisher – Google Scholar
Pathmanathan, S. G., Samat, N. A. & Mohamed, R. (2009). “Antimicrobial Susceptibility of Clinical Isolates of P. aeruginosa from a Malaysian Hospital,” Malaysian Journal of Medical Sciences, 16 (2) 146-152
Publisher
Ranjbar, R., Owlia, P., Saderi, H., Bameri, Z., Izadi, M., Jonaidi, N. & Morovvati, S. (2007). “Isolation of Clinical Strains of P. aeruginosa Harboring Different Plasmids,” Pak J Biol Sci, 10(17) 3020-2.
Publisher – Google Scholar
Sambrook, J. & Russell, D. W. (2001). “Molecular Cloning: A Laboratory Manual,” 3rd ed., New York: Cold Spring Harbor Laboratory Pres. 1.32-1.44.
Publisher – Google Scholar