Introduction
The first pandemic of the 21st century began in March of 2009 and was caused by new influenza Ð(H1N1)pdm09 virus strain of swine origin(CDC, 2010; Yatsyshina, 2011). This virus drew increased attention due to possible emergence of disastrous influenza pandemic comparable to Spanish influenza of 1918 having taken the lives of about 0.2— 8% infected people from different countries (up to50 million people worldwide)(Murray, 2006).
Influenza a (H1N1) pdm09 virus is thought to have occurred in Mexico in the middle of February, 2009, however, exact place and conditions of its origin are still unclear (Fraser, 2009). New A (H1N1) pdm09 virus probably emerged because of two genotypes (American and Eurasian) reassortment of swine influenza A (H1N1) virus (Neumann, 2009; Lvov, 2010) and therefore may be re-ferred as “quadruple” reassortant. High speed of epidemic development during March and April of 2009 affirmed that new antigenic influenza virus variant entered human population, crossed species barrier and was highly contagious(Baz, 2007; WHO, 2010; Yatsyshina, 2011).
Pandemic influenza viruses isolated in Russia from May 2009 to April 2010 were highly homologous to each other and to isolates from abroad, including references train Ð/California/04/2009(H1N1) pdm09 (Yat-syshina, 2011; Ilyicheva, 2011). Studies car-ried out in Russia revealed that during the 2009— 2010 season influenza had monoetio-logical nature that is typically in the pandemic period (Chan, 2010).
Influenza Ð (H1N1) pdm09 morbidity in Rus-sia can be divided into the following periods: pre-pandemic (April 27— June 11, 2009), initial pandemic (June 12 —August 15, 2009), and pandemic (August 15—December 1, 2009) (McCauley, 2012). During the pre-pandemic period the first case of human infection was identified in a Russian having returned from New Yorkto Moscow on May 21, 2009 (Lvov, 2010). Monitoring data of pandemic influenza virus in Ural, Siberian, and the Far Eastern Federal Districts of Russia showed that the first cases of Ð (H1N1) pdm09 in the Far East was detected 2—2.5 months later in comparison with Russian European part, and large increase occurred3 months later (Shchelkanov, 2010). From May 2009 to March 2010 samples from 862 patients suspected of having pandemic influenza were received from different regions of Russia by SRC VB “Vector”. Eighty nine strains Ð (H1N1) pdm09 (Agafonov, 2011) were isolated from those samples, including 56 isolates from residents of Siberian and the Far East regions (Ilyicheva, 2011). The total number of detected cases of infection according to The Center of Influenza Ecology and Epidemiology (Ivanovsky Institute of Virology, Moscow) reached 3053 by the beginning of December, 2009 (Chan, 2010).
On August 10, 2010 WHO declared cancella-tion of the 6th threat level for the world and the beginning of the post-pandemic period (WHO, 2010) when influenza Ð (H1N1) pdm09 virus circulation continued after changing into seasonal infection.
This study objectives were to compare bio-logical characteristics of influenza Ð (H1N1) pdm09 virus strains having circulated in the pandemic of 2009 and in the post-pandemic period in Russian Asia.
Materials and Methods
In the season of 2009—2010 in Russian Asia we isolated 56 pandemic influenza Ð(H1N1)pdm09 virus strains (Ilyicheva, 2011). Antigenicity and biomolecular features were similar for all isolates. In our study we used strain A/Novosibirsk/13/2009 (H1N1) pdm09. During the season of 2010—2011 we isolated 41 influenza Ð (H1N1) pdm09 virus strains (Sobolev, 2012). Antigenicity and biomolecular features were slightly different for strains within one group. In the study we used strain Ð/Novosibirsk/KSH/2011 (H1N1) pdm09 isolated at the very beginning of influenza epidemic in Western Siberia in January, 2011.
Viruses
- A/Novosibirsk/13/2009 (H1N1) pdm09 (N13-2009) (GU211227.1, GU211229.1, GU211231.1, GU211233.1, GU211228.1, GU211230.1, GU211234.1)
- Second passage with virus titer 6.30±0.24 lgTCID50/ml) isolated in September, 2009 from 28-years-old woman having returned from abroad. The woman recovered.
- Ð/Novosibirsk/KSH/2011 (H1N1) pdm09 (KSH-2011) (JQ041360.1, JQ041358.1, JQ041356.1, JQ041354.1, JQ041361.1, JQ041359.1, JQ041357.1, JQ041355.1) (second passage with virus titer 6.8±0.17 lgTCID50/ml) isolated in January, 2011 from 27-years-old man who had not been abroad. The man re-covered.
Animals. Eight-weeks-old BALB/c mice in the amount of 55 pieces were used in the experiment. Mice wereintranasally infected with 50 mcla 50% mouse infectious dose (MID50) of N13-2009 – 3,8lgMID50/ml and ofKSH-2011— 3,6 lgMID50/ml experienced weight loss. Animals were monitored for 15 days.
All manipulations with laboratory animals were performed in enhanced biosafety level 3 (BSL3) containment laboratories at the SRC VB “Vector” in accordance with the research protocol approved by the Bioethical Commit-tee of VECTOR.
Preparation of Lung Tissue Suspension. To detect virus concentration in lung tissue of infected animals 100mg of lung tissue were homogenated in porcelain jar with following addition of 9ml3 of Hanks’ solution with anti-biotics of the gentamicin and benzylpenicillin. The suspension was clarified by centrifu-gation.
Cells. Determination of virus titer was carried out in Madin-Darby canine kidney (MDCK) cell culture. MDCK cells were cultivated in Dulbecco’s Modified Eagle Medium(DMEM) (BioloT, Saint-Petersburg) with addition of 10% cow fetal serum (“HyClone”, Erembodegem, Belgium), 100 units per ml of benzylpenicillin in the atmosphere containing 5% of CO2 at 37ºÐ¡.Virus cytopathic effect was registered every day. Hemagglutination reaction was positive in each well where cytopathic effect was registered.
Antibody-Mediated Methods. Hemagglutina-tion inhibition (HI) and microneutralization (MN) assays were carried out in compliance with protocols developed by the Influenza Division at the Centers for Disease Control and Prevention (CDC), Atlanta, USA(WHO, 2011).
MID50 and TCID50 values were calculated by the Reed and Muench method (Reed, 1938).
Reference Sera. We used reference serum to virus Ð(H1N1)pdm09 kindly furnished by WHO Collaborating Centre for influenza (CDC, Atlanta, USA).
Strain titration was conducted in the HI assay using standard practice recommended by WHO with diagnostic serum to influenza A (H1N1) pdm09 virus and goose red blood cells. Real time polymerase chain reaction (PCR) was also carried out to subtype in-fluenza A viruses and differentiate influenza Ð (H1N1) pdm09 virus.
Genome Sequencing. RNA isolation was car-ried out using standard practice with “A set of reagents for the detection of swine in-fluenza A/H1 using PCR with hybridization-fluorescence detection “AmpliSens® Influenza virus A/H1-swine-FL” (Ilyicheva, 2011). PCR amplification and following sequencing were carried out using specific primerpairs recommended by WHO for work with in-fluenza Ð (H1N1) pdm09 viruses (WHO, 2009).
The analysis of amino-acid changes in hemag-glutinin gene (coding part using numeration typical for H1) and neuraminidase (coding part) was carried out with the program BioEdit ver.7.0.9.0.
Phylogenetic analysis of gene sequences was performed with the method of neighbor joining with the program MEGA 5.1. To evaluate reliability bootstrap test (1000 replications) was used. When building phylogenetic trees sequences from GISAID and GenBank data-bases were used.
Light Microscopy. Excised tissues of the lung of euthanized mice by dislocation of the cer-vical vertebrae at 1, 3, 6, and 10 days post infection were preserved in 10% phosphate-buffered formalin. Tissues were then processed for paraffin embedding and cut into 5-μm-thick sections. The section from each tissue sample was stained using a standard hematoxylin-and-eosin procedure. Light-optical study and microphotography were carried out using Axioskop 40 (Zeiss, Germany) microscope.
Results
We investigated virulent and immunogenic features of influenza virus strains N13-2009 and KSH-2011 in a mouse model. When mice were infected with virus strains symptoms of disease in both groups were registered since the first day. Disease was accompanied by conjunctivitis, decreased activity, decrease of body weight, breathlessness. The virustiters in lung were registered up to 6 days (Table 1).
Table 1: Virologic Values
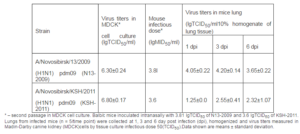
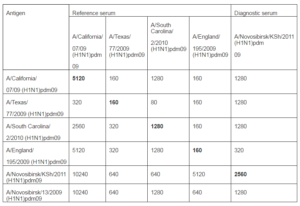
There were no fatal cases during all observa-tion time. Our results well correspond to previously obtained data (lyushina, 2010; Xu, 2011; Sakabea, 2011). Thus, it was shown that during infection of mice with viruses isolated from people animals deaths occurred only after virus adaptation by multiple passages in lung of animals. Antigenicity of strains were described in HI assay with diagnostic sera: reference serum Ð/California/07/2009(H1N1)pdm09 obtained from WHO Collaborating Centre for influenza (CDC, Atlanta, USA) and rabbit antiserum Ð/Novosibirsk/KSH/2011 (H1N1) pdm09 that we received using standard practice (Table 2).
Table 2: Antigenicity of Influenza Ð (H1N1) pdm09 Virus Strains in HI Assay with Reference Sera and Goose Red Blood Cells
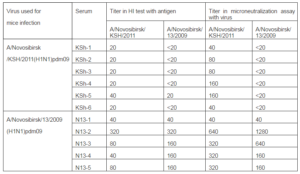
On the 15th day blood of animals that had illness was collected and sera were prepared. Sera were analyzed using HI and microneu-tralization assays with viruses used for infec-tion of mice. Results are shown in Table 3. As it seen from data of table mice infection with strain A/Novosibirsk/13/2009 (H1N1) pdm09 resulted in expressed humoral re-sponse: four sera of five have antibody titers to both influenza A (H1N1) pdm09 virus strains with higher values compared to thre-shold level (40 in HI assay and 80 in MN as-say). In sera from mice infected with strain Ð/Novosibirsk/KSH/2011(H1N1) pdm09 antibodies were detected only to “their own” strain and only using microneutralization assay.
Table 3: Hemagglutination Inhibitionand Microneutralization Assays with Serum from Mice Infected with Strains A/Novosibirsk/13/2009(H1N1) pdm09 and A/Novosibirsk/KSh/2011(H1N1) pdm09
We detected nucleotide sequence of hemagglutin in (HA) and neuraminidase (NA) genes and protein amino-acid sequence of influenza virus strains under study. Also was carried out comparative analysis between amino-acid sequences in HA and NA proteins of studied strains and proteins of one of the first influenza virus strains A/Mexico/InDRE4487/2009 (H1N1) pdm09 and the vaccine strain Ð/California/07/2009 pdm09 (Table 4). Strain N13-2009 proteins slightly differed from HA and NA of compared strains. Thus, amino-acid changes were detected in: antigenic site of hemagglutinin (Ca) I166T, S203T, R205K and in neuraminidase — V106I, N248D.Strain KSh-2011 had significantly more amino-acid changes: in subunit HA1— S143G, A197T, K211N, A215E, including changes in antigenic sites— (Ca) S203T, (Cb) L70P, (Sb) S185T; in subunit HA2— Е374К, S451N; and in neuraminidase — V106I, N248D.As a result, in the case of influenza Ð (Ð1N1) pdm09 virus isolated in the post-pandemic period, 2011, concentration of amino-acid changes was registered in HA gene compared to influenza virus circulated during the pandemic season of 2009, that was caused by virus antigenic drift under the influence of herd immunity.
Table 4: Amino-Acid Changes in HA and NA of Strains A/Novosibirsk/13/2009 (H1N1) pdm09 and A/Novosibirsk/KSH/2011(H1N1) pdm09
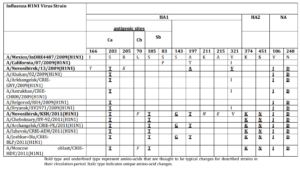
Bold type and underlined type represent amino-acids that are thought to be typical changes for described strains in their circulation period. Italic type indicates unique amino-acid changes.
Nucleotide sequences comparison of HA and NA of studied strains with relevant sequences of other strains from GISAID and GenBank databases helped us carry out phylogenetic analysis and build phylogenetic trees (Fig. 1, 2). Sequences were chosen taking into account the division into genotypes offered in the report of WHO Collaborating Centre for Reference and Research on Influenza in London (McCauley, 2012). According to this document 8 different genetic clades of HA and NA are distinguished on the base of amino-acid changes emerged during virus circulation in human population in 2009—2011.
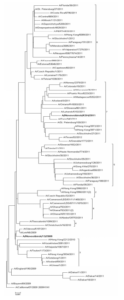
Fig. 1: HA Phylogenetic Tree
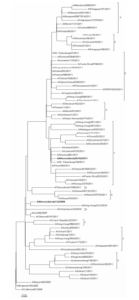
Fig. 2: NA Phylogenetic Tree
Phylogenetic features of HA and NA genes of the studied strain N13-2009 (bold type in the figure), isolated in 2009, are very similar to those of virus genes isolated in Ukraine and Hong Kong. Moreover, these viruses are not separated into special genetic clade and remain very similar to the vaccine strain A/California/07/2009. At the same time HA and NA genes of virus that we isolated in 2011 (bold type in the figure) belong to the seventh genetic clade that contains viruses circulating in 2011—2012 worldwide. Phylogenetic analysis revealed that strains under study belong to different genetic clades because of antigenic drift within virus population (Palese, 2011).
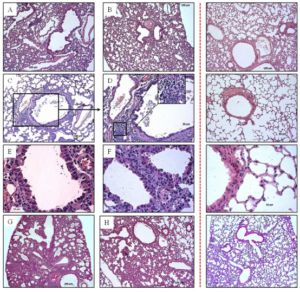
Fig. 3: Pathological Changes in Lung of Mice Infected with Influenza Ð (H1N1) pdm09 Virus
Lung of mice infected with strain A/Novosibirsk/13/2009: (A) 1 d. p. i.; (C—E) 6 d. p. i. (cellular epithelium desquamation, lymphocytic infiltration); (G) 10 d. p. i. (interstitial pneumonia). Lung of mice infected with strain A/Novosibirsk/KSh/2011: (B) 1 d. p. i.; (F) 6 d. p. i.; (H) 10 d. p. i. Control is shown behind the red dotted line.
Light-Optical Study
- Comparative analysis of pathological changes in lung tissue caused by infec-tion with strains N13-2009 and KSH-2011
On the first day post infection there were no significant changes. However, in lung tissue of mice infected with strain of 2009 we registered small concentration of erythro-cytes in alveoli, spasm of minute vessels and small in duration of surrounding respiratory tissue (Fig. 3 A, B).
On the third day after infection inflammatory process began in bronchial epithelium in both groups. Hygroscopic dystrophy of lung epithelium developed. However, in lung tissue of mice infected with isolates of 2009 pathologic process was visible not only in large and medium bronchi, but also in bronchial tubes. This points, in turn, at generalization of infection and viral particles invasion into lower respiratory tract.
On the sixth day post infection respiratory tissue airway obstruction was observed. Hypersecretion of mucin-producing cells of bronchioles was registered. Cell debris, multinucleated cells and mucus were visible in bronchi lumen (Fig. 3 C, E, F). We registered the focuses of leucocytic infiltra-tion comprising monocytes, lymphocytes and neutrophiles located in interalveolar septums, focuses of atelectasis and perivas-cularly. In interstitial tissue surrounding airways and alveolar sacs edema and multi-lobular necrosis were found. Plasmorrhagia were observed not only on blood vessel walls, but also in interstitium.
On the tenth day post infection we observed marked inflammatory changes in lung tissue of animals experimentally infected with strain N13-2009 (Fig. 3 G). Parenchyma airway obstruction dramatically decreased by type of atelectases and emphysematously dilatated alveoli. In the group of mice infected with strain KSh-2011 together with destruction processes in alveolar tissue we could discern the beginning of reparative processes that was confirmed by the presence of fibroblasts and collagen fibers (Fig. 3 H). Submucous layer edema was reduced.
Discussion
During this study we carried out compara-tive analysis of influenza Ð (H1N1) pdm09 virus strains isolated in Russian Asia in the pandemic of 2009 and in the post-pandemic period of 2011.Strain N13-2009 was isolated in Russia at the beginning of the pandemic period from 28-years-old woman having returned from Canada. She recovered. Another strain KSh-2011 was isolated in Russia in post-pandemic period (January, 2011) from 27-years-old man who had not been abroad. He recovered, too.
Comparison of virological characteristics of these strains revealed that both strains actively replicated in mice lung without previous adaptation. In case of strain N13-2009 virus titer accumulation in lung tissue was registered from 1 to 6 days similarly to strain KSh-2011. Though, on the first day we detected higher values of virus titers in lung in strains of 2009.
From all amino-acid changes in antigenic site Ca emerged in 2009 we detected that only change S203T remained and became one of the typical mutation among strains isolated in Russia in 2011. Amino-acid changes typical for all influenza Ð (Ð1N1) pdm09 viruses isolated in 2011 are the following: in subunit HA1— S143G, A197T, including S185T (Sb) in antigenic site; in HA2 — E374K, S451K; in NA gene— V106I, N248D (WHO, 2011; Baz, 2001; Agafonov, 2011). According to Kiselev (2011), replacements of the V106I, N248D in neuraminidase showed the immune pressure on the virus circulating. Two changes were detected in subunit HA2 of influenza virus isolated in 2011, including E374K originating at the beginning of 2009 pandemic. According to Kao et al (2012), it is that virus with this adaptive mutation that caused the peak of influenza morbidity in 2009—2010. The above listed changes in the structure of surface proteins of influenza viruses chosen for our study are the evidence of permanent antigenic drift in virus population causing change of genetic clades. We suppose that twin mutations of amino-acid changes in hemagglutininsuch is S185T, S143G, A197T, E374K and S451N effect on reducing virulent features.
Light-optical analysis of organs of mice in-fected with strains N13-2009 and KSh-2011 revealed pathological changes in lung tissue. Morphological study confirmed virological data indicating active replication in lung tissue of strains N13-2009 and KSH-2011. We detected that acute infectious disease— interstitial pneumonia— developed on the sixth day. Inflammatory process began in epithelial tissue of large and medium bronchi and then moved to interstitial tissue. Although both groups had microfocal and predominantly peribronchial interstitial pneumonia, in a whole inflammatory process was more severe when mice were infected with the strain of 2009. We suppose that mutation accumulation in genotype of pandemic influenza virus led to decrease of its pathogenicity by 2011 that had an impact on the course of inflammatory process in tissue of lung as the target organ.
Our research supported by date of the Fed-eral State Budgetary Institution «Research Influenza Institute» Ministry of Health and Social Development of Russia (Karpova, 2011). The epidemic 2011, in contrast to the pandemic period 2009, was lower intensity of involvement in the age groups population of the cities; for the duration of the epidemic; the incidence in the period of the epidemic of the whole population. Decreased hospitalization rate from the number of patients with influenza and acute respiratory viral infections and the percentage of deaths. However, the data presented Rodrígueza and others (2011) indicate significant differences in the post-pandemic period in 2011 between Russia and Spain. According to them, in patients admitted to the intensive care unit in the post-pandemic period of 2011 had higher frequency of severe comorbidities, severity of illness and increased mortality when compared with those observed in the 2009 pandemic outbreak.
In summary, we showed that pandemic influenza virus strain N13-2009 is more virulent and is able to cause more severe pathological processes in lung tissues com-pared to strain KSH-2011 isolated in the post-pandemic period. It seems that during influenza A(H1N1)pdm09 virus circulation from 2009 to 2011 in the structure of surface proteins mutations occurred, that caused change of genetic clade on phylogenetic tree and decrease of virulent features and presumably immunogenicity in virus biology.
Acknowledgements
The authors thank Dr. Starkova E.V. for par-ticipating in the discussion of the histologi-cal results. This work was supported: by The Ministry of education and science of Russia, projectsâ„– 14.B37.21.1927 and â„– 16.522.12.2011; by the U.S.D.A., Agricultural Research Service, under agreement â„– 58-0210-2-040F. The studies were performed using equipment access center FSBI SCCEM SB RAMS “Modern Optical System”.
We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database on which this research is based (see Table 2, Table 4, Figer 1 and Figer 2). All submitters of data may be contacted directly via the GISAID website www.gisaid.org
References
Agafonov, A. P., Shestopalov, A. M., Shikov, A. N., Demina, O. K., Durymanov, A. G., Ser-geev, A. A., Agafonova, P. A., Vinokurava, A. V., Sharshov, K. A., Berillo, S. A., Skarnovich, M. O., Sementsova, A. O., Ternovoi, B. A., Malkova, E. M., Stavsii, E. A. & Drozdov, I. G. (2011). ‘Studies of Properties of Pandemic Influenza Virus A/Ð1N1 Circulated in Russian Federation in 2009-2010,’ Zhurnal mikrobiologii, epidemiologii, i immunobiologii, (4) 24—27.
Baz, M., Abed, Y. & Boivin, G. (2007). “Characterization of Drug-Resistant Recombinant Influenza A/H1N1 Viruses Selected in Vitro with Peramivir and Zanamivir,” Antiviral research, 74 (2) 159—162.
Publisher – Google Scholar
Centers for Disease Control and Prevention (CDC). (2010). “The 2009 H1N1 Pandemic: Summary Highlights, April 2009— April 2010,” CDC-INFO. [Online], [Retrieved April 8, 2013], http://www.cdc.gov/h1n1flu/cdcresponse.htm
Publisher
Chan, M. (2010). ‘H1N1 in Post-Pandemic Period,’ WHO Media Center. [Online], [Retrieved April 8, 2013], http://www.who.int/mediacentre/news/state-ments/2010/h1n1_vpc_20100810/en/index.html
Google Scholar
Fraser, C., Donnelly, C. A., Cauchemez, S., Hanage, W. P., Van Kerkhove, M. D., Hol-linqsworth, T. D., Griffin, J., Baggaley, R. F., Jenkins, H. E., Lyons, E. J., Jombart, T., Hins-ley, W. R.,Grassly, N. C.,Balloux, F., Ghani, A. C., Ferguson, N. M., Rambaut, A., Pybus, O. G., Lopez-Gatell, H., Alpuche-Aranda, C. M., Chapela, I. B., Zavala, E. P., Guevara, D. M., Checchi, F., Garcia, E., Hugonnet, S. & Roth C.; WHO Rapid Pandemic Assessment Collaboration. (2009). “Pandemic Potential of a Strain of Influenza A (H1N1): Early Findings,” Science, 324 (5934) 1557—1561.
Publisher – Google Scholar
Ilyicheva, T., Susloparov, I., Durymanov, A., Romanovskaya, A., Sharshov, K., Kurskaya, O., Ignashkina, M. & Shestopalov, A. (2011). “Influenza A/H1N1pdm Virus in Russian Asia in 2009— 2010,” Infection, Genetics and Evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases, 11 (8) 2107—2112.
Publisher – Google Scholar
Ilyushina, N. A., Khalenkov, A. M., Seiler, J. P., Forrest, H. L., Bovin, N. V., Marjuki, H., Barman, S., Webster, R. G. & Webby, R. J. (2010). “Adaptation of Pandemic H1N1 Influenza Viruses in Mice,” Journal of virology 84 (17) 8607—8616.
Publisher – Google Scholar
Kao, C. L., Chan, T. C., Tsai, C. H., Chu, K. Y., Chuang, S. F., Lee, C. C., Li, Z. R., Wu, K. W., Chang, L. Y., Shen, Y. H., Huang, L. M., Lee, P. I., Yang, C., Compans, R., Rouse, B. T. & King, C. C.(2012). “Emerged HA and NA Mutants of the Pandemic Influenza H1N1 Vi-Ruses with Increasing Epidemiological Significance in Taipei and Kaohsiung, Taiwan, 2009-10,” PLoS One, 7 (2) e31162.
Publisher – Google Scholar
Karpova, L. S., Burtseva, E. I., Popovtseva, N. M. & Stoliarova, T. P. (2011). ‘Comparison of Influenza Epidemics in Russia 2009 and 2011, Caused by Pandemic Influenza A (H1N1),’ Epidemiology and vaccination, 5 (60) 6-15.
Kiselev, O. (2011). ‘Genome of Pandemic Influenza A/H1N1v-2009/10 Virus,’ Dmitreyd Grafik Group, St-Petersburg — Moskow, Russia.
Latsyshina, S. B., Minenko, A. N., Voloshina, P. V., Kudriavtseva, A. V., Praded, M. N.,Braslavskaia, S. I., Shipulin, G. A., Maleev, V. V. & Pokrovskii, V. I. (2011). “Molecular Characterization of Pandemic Influenza Viruses A/H1N1 (sw2009) Isolation in Russia in 2009-2010,” Zhurnal mikrobiologii, epidemiologii, i immunobiologii, (1) 26—34.
Publisher
Lvov, D. K., Burtseva, E. I., Prilipov, A. G., Bazarova, M. V., Kolobukhina, L. V., Merkulova, L. N., Malyshev, N. A., Deriabin, P. G., Fediakina, I. T., Sadykova, G. K., Usachev, E. V., Shchelkanov, M.Iu., Shevchenko, E. S.,Trushakova, S. V., Ivanova, V. T., Beliakova, N. V., Oskerko, T. A. & Ali-per, T. I. (2009). “The 24 May, 2009 Isolation of the First A/IIV-Moscow/01/2009 (H1N1) swl Strain Similar to Swine A (H1N1) Influenza Virus from the First Moscow Case Detected on May 21, 2009, and its Deposit in the State Collection of Viruses,” Voprosy virusologii, 54 (5) 10—14.
Publisher – Google Scholar
Lvov, D. K., Burtseva, E. I., Shelkanov, M. Yu. et al. (2010). ‘Spread of New Pandemic Influenza A (Ð1N1) v Virus in Russia,’ Voprosy virusologii, 55 (3) 4—9.
McCauley, J., Daniels, R., Lin, Y., Zheng, X., Gregory, V., Whittaker, L., Cattle, N., Halai, C. & Cross, K. (2012). “Report Prepared for the WHO Annual Consultation on the Composition of Influenza Vaccine for the Northern Hemisphere,” IOP Publishing WHO influenza centre. [Online], [Retrieved April 8, 2013], http://www.nimr.mrc.ac.uk/documents/about/interim-report-feb-2012.pdf
Publisher
Murray, C. J. L., Lopez, A. D., Chin, B., Feehan, D. & Hill, K. H. (2006). “Estimation of Potential Global Pandemic Influenza Mortality on the Basis of Vital Registry Data from the 1918-20 Pandemic: A Quantitative Analysis,“ Lancet, 368 (9554)2211—2218.
Publisher – Google Scholar
Neumann, G., Noda, T. & Kawaoka, Y. (2009). “Emergence and Pandemic Potential of Swine-Origin H1N1 Influenza Virus,” Nature, 459 (7249) 931—939.
Publisher – Google Scholar
Palese, P. & Wang, T. T. (2011). “Why Do Influenza Virus Subtypes Die out? A Hypothesis,” mBio, 2 (5) e00150-11.
Publisher – Google Scholar
Reed, L. J. & Muench, H. (1938). ‘A Simple Method of Estimating Fifty Percent Endpoints,’ American Journal of Hygiene, (27) 493—497.
Google Scholar
Rodrígueza, A., Martin-Loechesa, I., Bona-streb, J., Olaechea, P., Alvarez-Lerma, F., Zaragoza, R., Guerrero, J., Blanco, J., Gordo, F., Pozo, F., Lorente, J., Carratalá, J., Cordero, M., Rello, J., Esteban, A. & León, C.; SEMICYUC-CIBERES-REIPI working group. (2011). “First Influenza Season after the 2009 Pandemic Influenza: Report of the First 300 ICU Admissions in Spain,” Medicina intensiva, 35 (4) 208–216.
Publisher – Google Scholar
Sakabea, S., Ozawab, M., Takanoa, R., Iwas-tuki-Horimoto, K. & Kawaoka, Y.(2011). “Mutations in PA, NP, and HA of a Pandemic (H1N1) 2009 Influenza Virus Contribute to its Adaptation to Mice,” Virus research, 158 (1-2) 124—129.
Publisher – Google Scholar
Shchelkanov, M. Iu., L’vov, D. N., Fediakina, I. T., Shchelkanov M. Iu., L’vov, D. N., Fediakina, I. T., Baranov, N. I., Gorelikov, V. N., Reznik, V. Ia., Zdanovskaia, N. I., Pukhovskaia, N. M., Avdoshina, L. N., Shapiro, N. I., Snetkova, I. P., Kozhan, V. N., Iarovenko, G. M., Kalaeva, E. E., Gromova, M. A., ElovskiÄ, O. V., Eremeeva, Iu. V., Dovgal’, M. A., Kuchenkov, A. A., Anan’ev, V. Iu., Burtnik, V. I., Ivanov, L. I., Garbuz, Iu. A., Podolianko, I. A., Grigor’ev, S. N., Proshina, E. S., Samokhvalov, E. I., Al’khovskiÄ, S. V., Burtseva, E. I., Prilipov, A. G., Abbasova, E. I., Mironenko, E. S., Kolobukhina, L. V., Deriabin, P. G., Ott, V. A., Maslov, D. V., Ianovich, V. A. & L’vov, D. K.(2010). “Trends in the Spread of Pandemic Influenza A(H1N1) swl in the Far East in 2009,” Voprosy virusologii, 55 (3) 10—15.
Publisher – Google Scholar
Sobolev, I., Kurskaya, O., Susloparov, I., Ilyi-cheva, T. & Shestopalov, A. (2012). “Molecular Genetic Analysis of Influenza A/H3N2 Virus Strains Isolated in Western Siberia in the 2010-2011 Epidemic Season,” Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases, 12 (8) 1694-1698.
Publisher – Google Scholar
World Health Organization. (2009). “CDC Protocol of Realtime RTPCR for Influenza A (H1N1),” [Online], [Retrieved April 8, 2013], http://who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html
Publisher
World Health Organization. (2010). “H1N1 in Post-Pandemic Period,” [Online], [Retrieved April 8, 2013], http://www.who.int/mediacentre/news/state-ments/2010/h1n1_vpc_20100810/en/index.html
Google Scholar
World Health Organization. (2010). “Pandemic (H1N1) 2009— Update 112,” [Online], [Retrieved April 8, 2013],http://www.who.int/csr/don/2010_08_06/en/index.html
Publisher
World Health Organization. (2011). “Manual for The Laboratory Diagnosis and Virological Surveillance of Influenza,” OIP Publishing Whqlibdoc. [Online], [Retrieved April 8, 2013], http://www.who.int/influenza/gisrs_laborato-ry/manual_diagnosis_surveillance_influenza/en/index.html
Google Scholar
Xu, L., Bao, L., Li, F., Ma, Y., Zhou, J., Xu, Y., Deng, W., Zhan, L., Zhu, H., Ma, C., Shu, Y. & Qin, C. (2011). “Adaption of Seasonal H1N1 Influenza Virus in Mice,” PLoS One, 6 (12) e28901.
Publisher – Google Scholar