Introduction
Blood stream infections (BSIs) are a potentially life-threatening condition with a case fatality rate of 20-50% and among the most common health-care associated infections reported by Gupta et al. (2010) and Khawaja et al. (2013). Changing epidemiology and antimicrobial susceptibility among blood stream pathogens is a significant problem worldwide with severe consequences including increased cost of care, morbidity and mortality highlighted by Gupta et al. (2010) and Pourakbari et al. (2012). BSIs are affecting over 200,000 individuals annually with a mortality rate of 32% in the United States alone reported by Pourakbari et al. (2012). Antimicrobial resistance among blood stream pathogens is a significant problem around the globe. This is especially true in countries like Pakistan, where antibiotics are widely used and considerable resistance has been reported from all over the country (Akhtar, 2010 and Khawaja et al., 2013). The increasing resistance to many antibiotic compounds results in a reduction in therapeutic options suggested by Pourakbari et al. (2012). Hence detection of BSIs must be a priority in all clinical settings. The blood culture remains a gold standard in the diagnosis of BSIs.
Selecting appropriate antimicrobial for treating BSI is multifaceted; including possible cause and source of infection, in vitro activity of the drug according to microbiological susceptibility testing results, pharmacokinetics and adverse effects of the drug. However, before considering these aspects, the choice of antibiotic mainly relies on knowledge of the pathogen likely to be involved. For the benefit of patient, empirical therapy often has to be initiated without exact knowledge of the causative organism and its antimicrobial susceptibility profile, confirmed by Assadian et al. (2009). Monitoring and analyzing the antimicrobial susceptibility patterns of most frequently isolated microorganisms according to local epidemiology helps clinicians to choose effective empirical therapies and develop rational prescription policy for antibiotics. Therefore, analysis of occurrence of pathogens and their cumulative susceptibility profiles is widely used in hospital settings as suggested by Assadian et al. and Barati et al. (2009). A study conducted by Paul et al. (2010) highlighted that mortality was significantly higher among patients receiving inappropriate empirical therapy.
Different studies have been published regarding frequency and pattern of nosocomial infections in tertiary care settings but limited work has been reported regarding bacterial pathogens causing BSIs in our country in the last decade (Mumtaz et al. and Shaikh et al., 2008 and Latif et al., 2009). The objective of this study was to determine the pattern of pathogens and their antimicrobial susceptibility from blood culture specimens in a tertiary care setting to facilitate the formulation of antibiotic policy for empirical treatment.
Materials and Methods
This study was carried out at the Department of Microbiology, Armed Forces Institute of Pathology (AFIP) Rawalpindi, Pakistan. Permission from Institutional Ethical Committee & Institutional Review Board was taken. Sampling technique was non probability consecutive; all admitted patients with unexplained/undiagnosed fever, whose blood cultures specimens were sent to AFIP were included in the study. Contaminated, duplicate and repeat specimens from same patient were excluded from the study. Blood sample was collected aseptically from each patient before the start of antimicrobial therapy; in adults 8ml blood was added to each of brain heart infusion (BHI) broth (Oxoid, UK) and automated BACTEC 9050 (BD Diagnostics) culture bottles; and in children 3ml. Blood culture specimens from patients admitted in different wards and intensive care units of two 1000-bedded tertiary care referral hospitals at Rawalpindi, Urology and Bone marrow Transplant institutes were dealt with during the study. Manual and automated BACTEC 9050 (BD Diagnostics) blood culture systems were used for blood culture. For the manual method, BHI bottles were incubated at 37oC after shaking and sub-cultured as per standard protocol on blood and MacConkey’s agar (Oxoid, UK). Gram staining was performed on smears from the broth of positive culture bottles. Growth indicated by the automated system was sub-cultured on blood and MacConkey’s agar followed by colony morphology, Gram staining, motility testing, rapid tests like catalase, coagulase, oxidase and other requisite biochemical tests. For species identification of Gram positive organisms API Staph, API STREP, API Coryne (BioMérieux, France) were utilized and for Gram negative isolates API 20E, API 20NE (BioMérieux, France). Specimens yielding yeast were identified through colony morphology followed by speciation with Candida chromagar (Oxoid, UK) and API 20C AUX (BioMérieux, France). Disc diffusion technique (Kirby-Bauer) was used for antimicrobial susceptibility according to the guidelines of Clinical and Laboratory Standards Institute (CLSI 2012) and zones of inhibition were interpreted accordingly. Control strains were used as per requirement:ATCC 25923 Staphylococcus aureus, ATCC 27853 Pseudomonas aeruginosa, ATCC 25922 Escherichia coli, ATCC 19606 Acinetobacter baumannii, ATCC 10231 Candida albicans. The data was entered in SPSS (version 17) software. Descriptive statistics were calculated for both qualitative & quantitative variables. Qualitative variables like drugs’ susceptibility, frequency & percentages were calculated.
Results
Out of 2921 blood culture specimens received from different wards, 465 (16%) yielded growth of different microorganisms. The mean age of the patients was 24.03 + 20.22 SD; range 7 months to 75 years. The majority of the patients were males (72%); male to female ratio was 2.6:1. Out of these, 245 (53%) isolates were Gram positive, 209 (45%) were Gram negative while 11 (2%) isolates were yeast (Candida spp.). Staphylococcus spp. (208) was the leading pathogen in the list of Gram positive isolates, while Acinetobacter spp. (43), Pseudomonas spp. (43), E. coli (41) andSalmonella spp. (30) were the main pathogens amongst Gram negative isolates. Out of 208 Staphylococci, 180 (86.5%) were coagulase negative and 158 (76%) were methicillin resistant. Amongst Gram negative group, 115 (55%) isolates were members of enterobacteriaceae family, and E. coli (41) was the leading pathogen, while 94 (45%) were non fermenters (NF). Among these, Acinetobacter spp. and Pseudomonas aeruginosa (43 each) were the leading pathogens.
The detailed microbiology of pathogens isolated and the antimicrobial susceptibility of Gram positive and Gram negative organisms causing blood stream infections is shown in figure 1 & 2 respectively. Clinical outcome of patients from different clinical settings after antibiotic therapy following culture report is shown in table.
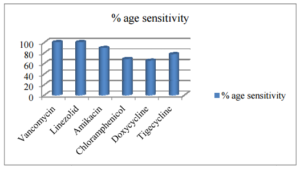
Figure 1. Susceptibility Pattern of Staphylococcus Spp. (N=208)
Drug susceptibility of Gram positive isolates to penicillin, cloxacillin, cephradine, amoxicillin+clavulanate and erythromycin was less than 20%.
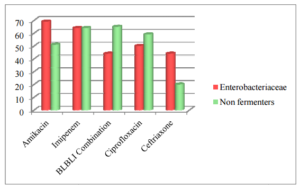
Figure 2. Susceptibility Pattern of Gram Negative Isolates (N=209) % Age Sensitivity
Drug susceptibility of Gram negative isolates to ampicillin, amoxicillin+clavulanate, gentamicin, cotrimoxazole, doxycycline, ceftazidime and cefepime was less than 40 %.
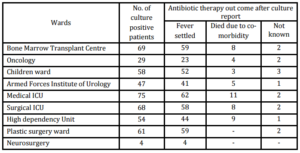
Table . Break Down of Patients from Different Wards and Outcome after Antibiotic Therapy Following Culture Report (n=465)
Discussion
Antimicrobial resistance is a well-known and ongoing problem in all bacterial pathogens. Blood stream infections are often due to strains that are resistant to a wider range of antimicrobial agents and are of particular concern in clinical practice reported by Gupta et al. (2010) and Alam et al. (2011). Over the last few years, research (Assadian et al., 2009; Moyo et al., 2010; Wang et al., 2012) showed that there has been an increase in Gram positive organisms especially coagulase negative staphylococci (CoNS) and Staphylococcus aureus as causative agents in causing BSI. This finding was elucidated partly by the increased use of intravascular devices, which can serve as portals of entry to the bloodstream. Similar trend was noted in our study. A study carried out in Saudi Arabia by Asghar, (2006) from septicemic patients showed that Gram positive isolates predominately S. aureus and CoNS accounted for about 57% of BSIs. Among Gram negative organisms, the majority of isolates were Pseudomonas spp., Acinetobacter spp. and E. coli. Our results were almost in concordance with these findings. Results of studies from Iran by Barati et al. (2009), Pourakbari et al. (2012) and Bangladesh by Darmstadt et al. (2009) were also in conformity with our findings as regards pathogens causing BSIs.
The organisms causing BSIs have shown higher resistance to ciprofloxacin and third generation cephalosporins; similar trend has been reported by an Indian study (Kollef 2008). The members of family enterobacteriaceae showed more resistance to third generation cephalosporins and β-lactam/β-lactamase inhibitor combinations. Studies from other parts of the world (Kollef 2008; Hashairi et al., 2011; Datta et al., 2012; Kenyon et al., 2012) also reported significant resistance to these groups of antibiotics for Gram negative rods. On the other hand, amikacin and imipenem yielded better activity against enterobacteriaceae family. In case of non fermenters, β-lactam/β-lactamase inhibitor combinations comprising of tazobactam/piperacillin, sulbactam/cefoperazone and sulbactam/piperacillin as well as carbapenems revealed better in vitro activity. These results are comparable to work done at other centres (Kollef, 2008; Hashairi et al., 2011; Datta et al., 2012; Kenyon et al., 2012; Chhina and Gupta, 2013; Tiwari and Kaur, 2010).
Methicillin resistance among staphylococci causing BSIs was alarmingly high in our setup and similar pattern was reported from a study carried out in Iran by Pourakbari et al. (2012). Methicillin resistance in our region was much more as compared to that reported from East Africa by Moyo et al. (2010) and India by Chhina and Gupta, (2013), where it was 23% and 41% respectively. This prompts us for meticulous antibiotic policies and strict implementation of hospital infection control measures.
Most of the isolates in our study were either from immune-compromised patients or serious patients in ICUs. Most of them fit in the category of hospital acquired infections. This also highlights the importance of prevailing organisms and their sensitivity pattern.
The rapid emergence of antibiotic resistance and its implications on empirical therapy point towards all clinical settings that they should have active ongoing microbiological surveillance. Such effort of monitoring the antibiotic-resistant pathogens would definitely benefit the current antimicrobial regimens especially in a resource poor country like ours. One of the factors for low yield of blood culture was over advice for blood cultures as these hospitals are providing free healthcare services. The observation in this study has been communicated to the concerned authorities for proper advice.The data on bacterial pathogens and antibiotic susceptibility, we believe, are an important pillar in our efforts at improving infection control practices by formulating policy for empirical treatment. We will continue the surveillance in future that will definitely improve novelty and robustness of data regarding the changing profile, if any, of pathogens and their susceptibility.
Conclusion
Gram positive organisms were predominant causing BSIs in our setup. Resistance to methicillin among Staphylococcusspp. & ceftriaxone resistance in Gram negative organisms are on the rise. Amikacin and tigecycline showed better in vitroefficacy against Gram positive as compared to Gram negative isolates, while susceptibility of carbapenems has decreased in both lactose fermenters as well as non fermenters. Vancomycin and linezolid yielded excellent in vitroactivity against Gram positive organisms.
References
1.Akhtar, N. (2010) “Hospital Acquired Infections in a Medical Intensive Care Unit,” Journal of the College of Physicians and Surgeons Pakistan, 20 (6) 386-90.
Publisher – Google Scholar
2.Alam, M. S. Pillai, P. K. Kapur, P. & Pillai, K. K. (2011) “Resistant Patterns of Bacteria Isolated from Bloodstream Infections at a University Hospital in Delhi,” J Pharm Bioallied Sci, 3 (4) 525-30.
Publisher – Google Scholar
3.Asghar, A. H. (2006) “Frequency and Antimicrobial Susceptibility Patterns of Bacterial Pathogens Isolated from Septicemic Patients in Makkah Hospitals,” Saudi Medical Journal, 27 (3) 361-7.
Publisher – Google Scholar
4.Assadian, O., Diab-Elschahawi, M., Makristathis, A., Blacky, A., Koller, W. & Adlassnig, K.-P. (2009) “Data Correction Pre-Processing for Electronically Stored Blood Culture Results: Implications on Microbial Spectrum Empiric Antibiotic Therapy,” MC Medical Informatics and Decision Making, 9:27. doi:10.1186/1472-6947-9-27.
Publisher – Google Scholar
5.Barati, M. Taher, M. T. Abasi, R. Zadeh, M. M. Barati, M. & Shamshiri, A. R. (2009) ‘Bacteriological Profile and Antimicrobial Resistance of Blood Culture Isolates,’ Iranian Journal of Clinical Infectious Diseases, 2 (4) 87-95.
6.Chhina, D. & Gupta, V. (2013) ‘Bacteriological Profile and Antimicrobial Susceptibility Pattern Of Blood Isolates from a Tertiary Care Hospital in North India,’ IJPRBS, 2 (2) 24-35.
Google Scholar
7.Clinical and Laboratory Standards Institute (2012). ‘Performance Standards for Antimicrobial Susceptibility Testing, 22nd Informational Supplement,’ M100-S22. Clinical and Laboratory Standards Institute (CLSI), Wayne, PA,2012.
8.Darmstadt, G. L. , Saha, S. K., Choi, Y., El. Arifeen, S., Ahmed, N. U. & Bari, S. et al. (2009) “Population-Based Incidence And Etiology of Community-Acquired Neonatal Bacteremia in Mirzapur, Bangladesh: An Observational Study,”The Journalof Infectious Diseases, 200 (6) 906-15.
Publisher – Google Scholar
9.Datta, S., Wattal, C., Goel, N., Oberoi, J. K., Raveendran, R. & Prasad, K. J. (2012) “A Ten Year Analysis of Multi-Drug Resistant Blood Stream Infections Caused by Escherichia Coli & Klebsiella Pneumoniae In A Tertiary Care Hospital,”Indian Journal of Medical Research, 135 (6) 907-12.
Publisher – Google Scholar
10.Gupta, A., Sharma, S., Arora, A. & Gupta, A. (2010) “Changing Trends of in Vitro Antimicrobial Resistance Patterns in Blood Isolates in a Tertiary Care Hospital over a Period of 4 Years,” Indian Journal of Medical Sciences 64 (11) 485-92.
Publisher – Google Scholar
11.Hashairi, F., Hasan, H., Azlan, K. & Deris, Z. Z. (2011) “An Eight-Year Review of Blood Culture and Susceptibility among Sepsis Cases in an Emergency Department in Northeastern Malaysia,” Tropical Biomedicine, 28 (3) 599-605.
Publisher – Google Scholar
12.Kenyon, C.R., Fatti, G., Schrueder, N., Bonorchis, K. & Meintjes, G. (2012) “The Value of Blood Culture Audits at Peripheral Hospitals,” South African Medical Journal, 102 (4) 224-5.
Publisher – Google Scholar
13.Khawaja, A., Zubairi, A. B., Durrani, F. K. & Zafar, A. (2013) “Etiology and Outcome of Severe Community Acquired Pneumonia in Immunocompetent Adults,” BMC Infectious Diseases , 13:94. doi: 10.1186/1471-2334-13-94.
Publisher – Google Scholar
14.Kollef, M. H. (2008) “Broad-spectrum Antimicrobials and the Treatment of Serious Bacterial Infections: Getting it Right up Front,” Clinical Infectious Diseases, 47 (1) 3-13.
Publisher – Google Scholar
15.Latif, S., Anwar, M. S. & Ahmad, I. (2009) “Bacterial Pathogens Responsible for Blood Stream Infection (BSI) and Pattern of Drug Resistance in a Tertiary Care Hospital of Lahore,” Biomedica, 25(2) 101-5.
Publisher – Google Scholar
16.Moyo, S., Aboud, S., Kasubi, M. & Maselle, S. Y. (2010) “Bacteria Isolated from Bloodstream Infections at a Tertiary Hospital in Dar Es Salaam, Tanzania–Antimicrobial Resistance of Isolates,” South African Medical Journal, 100 (12) 835-8.
Publisher – Google Scholar
17.Mumtaz, S., Ahmad, M., Aftab, I., Akhtar, N. & Hameed, A. (2008) “Frequency of Extended-Spectrum -Lactamases (ESBLs) and Blood Stream Infections,” Infectious Diseases Journal of Pakistan, 17 (2) 48-51.
Publisher – Google Scholar
18.Paul, M., Kariv, G., Goldberg, E., Raskin, M., Shaked, H. & Hazzan, R. et al. (2010) “Importance of Appropriate Empirical Antibiotic Therapy for Methicillin-resistant Staphylococcus Aureus Bacteraemia,” Journal Antimicrobial Chemotherapy, 65 (12) 2658-65.
Publisher – Google Scholar
19.Pourakbari, B., Sadr, A., Ashtiani, M. T., Mamishi, S., Dehghani, M. & Mahmoudi, S. et al. (2012) “Five-year Evaluation of the Antimicrobial Susceptibility Patterns of Bacteria Causing Bloodstream Infections in Iran,” The Journal of Infection in Developing Countries, 6 (2) 120-5.
Publisher – Google Scholar
20.Shaikh, J. M., Devrajani, B. R., Shah, S. Z. A., Akhund, T. & Bibi, I. (2008) “Frequency, Pattern and Etiology of Nosocomial Infection in Intensive Care Unit: An Experience at a Tertiary Care Hospital,” J Ayub Med Coll Abottabad, 20 (4) 37-40.
Publisher – Google Scholar
21.Tiwari, P. & Kaur, S. (2010) “Profile and Sensitivity Pattern of Bacteria Isolated from Various Cultures in a Tertiary Care Hospital in Delhi,” Indian Journal of Public Health, 54 (4) 213-5.
Publisher – Google Scholar
22.Wang, H.J., Shi, H., Zhou, W., Hu, Z. Q., Mu, L. Y. & Su, M. et al. (2012) “Common Pathogens and Clinical Characteristics of Neonatal Pneumonia,” Zhongguo Dang Dai Er Ke Za Zh, 14 (12) 898-902.
Publisher – Google Scholar