Introduction
Cancer of the uterine cervix is the second most common cancer among women worldwide (Parkin and Bray 2006), but it is the leading cancer in Indian women with an annual incidence of about 130,000 cases and 70,000-75,000 deaths (Das et al. 2008). Various clinico-epidemiological studies have established high-risk human papillomavirus (HR-HPV) as the major etiological agent in cervical carcinogenesis (Das et al. 2008). However, in 70—90% of HPV-infected individuals, the virus is naturally cleared. Only a fraction of these individuals, with persistent infection of HR-HPV, develop cervical cancer after a long latent period (Zur 2002). Thus, HPV infection alone is not sufficient, but together with immunological, environmental or genetic cofactors might mediate cervical carcinogenesis (Zur 2002; Kohaar et al. 2007). Among the host genetic factors, variations in effective host immune response may be an important determinant of persistence of HPV infection and susceptibility to cervical cancer.
Proinflammatory cytokines, Tumor Necrosis Factor α (TNF) and a related cytokine Lymphotoxin α (LTA) play a major role in the cell mediated immunity (CMI) of the host, thereby constituting important genetically determined host factors in cervical cancer. Both the genes are closely spaced and tandemly arranged in HLA class III region on chromosome 6p21.3. TNF and LTA bind to both TNF-RI and TNF-RII receptors and share many biological activities (Aggarwal 2003). The crucial role of TNF in carcinogenesis was established from the study on the knockout mouse model of TNF- receptor type 1 (Knight et al. 2000). In case of cancer of uterine cervix, it has been implicated in the control of HPV infection in several in vivo and in vitro studies (Bequet-Romero and Lopez-Ocejo 2000; Malejczyk et al. 1992; Tjiong et al. 2001). In addition, elevated plasma levels of TNF and LTA have been associated with various inflammatory, autoimmune, and malignant diseases (Beutler and Bazzoni 1998; Balkwill 2002). TNF expression levels may thus be a determinant of pathogenesis and disease progression in cancer (Moore et al. 1999). The expression is mostly regulated at the transcriptional level and interestingly, it is believed to be influenced by genetic polymorphisms within their genes (Wilson et al. 1997; Messer et al. 1991). Several of these SNPs have been associated with susceptibility to various infectious and non-infectious diseases including cancer (Kaluza et al. 2000; Wilson et al. 1995; Gonzalez et al. 2003; Ozaki et al. 2002).
Therefore, the present study has been designed to investigate the functional polymorphisms/ haplotypes in TNF-LTA locus for their role in the predisposition towards HPV mediated cervical cancer in a population-based case-control study. We also analyzed the influence of polymorphisms in TNF-LTA locus on the regulation of TNF and LTA production.
Materials and Methods
Subjects
Population
A total of 200 patients (Indo-Aryan ethnicity) comprising 50 cervical precancer (CIN 2/3) and 150 invasive cervical cancer cases were employed for the study. The patients were recruited from Safdarjung Hospitals, New Delhi, with histopathologically confirmed precancer/invasive carcinoma of uterine cervix. The patients had a mean age of 49.6 ± 11.3 yrs. The age (47± 12 yrs) and ethnicity matched control group consisting of 200 healthy women with no self or family history of any neoplastic disease were from out-patients of the Department of Gynaecology, Safdarjung hospital, New Delhi, who came for routine check-up. 5 ml blood samples were also collected from 85 cases (15 precancer and 70 invasive cancer) and 90 controls in EDTA coated vacutainers. Plasma was separated and stored at -70ËšC within an hour of collection for further analysis.
Written consent was obtained from all the participants and the study was carried out in accordance with the principles of Helsinki Declaration and was approved by the Ethics Committee of the Institute.
Samples and Processing
Genomic DNA Extraction
Genomic DNA was extracted from freshly collected tissue biopsies from patients and cervical scrapes from control subjects by standard method using proteinase K followed by phenol/chloroform/isopropanol treatment (Sambrook et al. 1989) for HPV screening and polymorphism studies.
HPV Screening
HPV diagnosis was performed by PCR amplification using consensus primers MY09 and MY11 (Das et al. 1992), and further typing was done by PCR using type specific primers for HPV 16 and HPV18 (Saiki et al. 1988). β-globin primers PC04 and GH20 which produce 268bp PCR products were used as an internal amplification control (Saiki et al. 1985). Positive and Negative controls were also taken with every round of amplification.
Analysis of TNFA and LTA Polymorphisms
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) approach was employed to genotype all the SNPs in TNF —LTA region. -238 G/A (rs361525) and -308 G/A (rs1800629) were genotyped by method employed by Jang et al (Jang et al. 2001), with some modification (Kohaar et al. 2007). While -857 C/T (rs1799724), -863 C/A (rs1800630) & -1031 T/C (rs1799964) were genotyped according to Skoog et al (Skoog et al. 1999).
In case of LTA (+252 A/G) locus, primers (Forward Primer- 5’-GCT TCG TGC TTT GGA CTA CC-3’; Reverse Primer-5’-AAG GTG AGC AGA GGG AGA CA-3’) were designed using the software PRIMER 3.0. The reaction conditions were: 94°C for 5 min; 30 cycles of 94°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec and final extension of 72°C for 5 min. For the RFLP analysis, 6µ1 (~100 ng) of the PCR products were digested at 37°C in a 20 µ1 reaction mixture containing 2.5 units with NcoI (Fast Digest, MBI Fermentas) to detect the +252 polymorphism. NcoI cleaves the amplified fragments containing G at +252 position producing two fragments of 196bp and 50bp. The RFLP analysis was performed on 10% native polyacrylamide gel.
We sequenced 20% of the samples randomly to validate the data generated by PCR-RFLP method. Sequencing reactions were performed according to the conventional dideoxy chain termination method using ABI PrismTM 310 Automated DNA Sequencer (Applied Biosystem, USA).
In-vitro Quantitative Determination of TNFA and LTA
Sandwich Enzyme Linked Immunosorbent Assay (ELISA) was employed for in-vitro quantitative determination of TNF α (Human TNF α ELISA; Diaclone; France) and LTA (Human LTA ELISA; Bender MedSystems; Austria) from the plasma according to the manufacturer’s instructions.
Statistical Analysis
The data analysis was performed using the computer software Statistical Package for the Social Sciences (SPSS) for Windows (version — 12.0). Chi-square test / Fisher’s Exact Test (for smaller numbers on subgroup analysis) was used to compare the distributions of TNFA-LTA polymorphisms between cases and controls. Haplotypes were constructed from genotypes of 6 polymorphic markers in TNF-LTA locus by using PHASE (http://linkage.rockfeller.edu). LD estimates were determined by Haploview (http://www.broad.mit.edu/mpg/haploview/). Kruskal Wallis and Mann Whitney U test were performed for quantitative parameters. Corrected P value (Pc) was obtained after applying Bonferroni’s correction. Pc values (two-tailed) of <0.05 were considered statistically significant.
Results
HPV Prevalence
In the studied population, about 87.0% (174/200) of cases and 2.5% (5/200) of normal healthy controls showed positivity for HPV DNA sequence. Out of the HPV positive cases, 98.17% (171/174) were infected with HPV type16 and rest 1.8% (3/164) were found to be positive for HPV type 18. While all the HPV positive healthy controls were found to be infected with HPV type 16. After the stratification of cases, it was found that 91.33% (137/150) of invasive cancer cases and 74.0% (37/50) of precancerous cases were HPV positive.
Polymorphisms in TNF-LTA Locus
Genotype frequencies for all polymorphisms were found to be in concordance with Hardy—Weinberg Equilibrium in both cases and controls (P > 0.05) (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) (Sasieni 1997). The distribution of TNF- LTA genotypes and haplotypes in both cases and controls is depicted in Tables 1 and 2. Supplementary Tables 1 and 2 show distribution frequencies of single nucleotide polymorphisms in TNFA and LTA with respect to HPV infection status.
A significant difference (Pc = 0.01) was observed with respect to TNFA —308 carrier A (GA/AA) genotype distribution between cases and controls. The association was also found to be highly significant (Pc = 0.01) in carcinoma cases in comparison to the control group but in pre-cancerous cases, it could not attain statistical significance (P = 0.11). Also, the variant homozygous genotype —308 AA was found only in case: 2.0% (1/50) in precancer and 3.33% (5/150) in cancer, but absent in controls. The —308 (G/A) TNFA promoter genotype with respect to HPV status revealed that “A” allele was significantly associated (Pc = 0.009) with HPV positive cases of cervical precancer and cancer.
Carrier genotype (CT/TT) distribution at -857 C/T locus revealed statistically significant association (P=0.011) for controls vs. total cases (precancer and invasive cancer) and controls vs. cancer group (P =0.0069). However, after applying correction for multiple testing, the significance was retained only for controls vs. cancer group (Pc for controls vs. cancer = 0.04; Pc for controls vs. total cases = 0.06). 4.0% (8/200) of the total cases (2.0%; 1/50 in precancer and 4.67% (7/150) in cancer group) and 6.5% (13/200) of controls were found to be polymorphic homozygous (TT).
In case of -1031 T/C SNP (rs1799964), a significant association was established for carrier genotype (-1031TC/CC) distribution between HPV positive cases vs. controls (P=0.012) and HPV positive cases vs. HPV negative cases (P=0.001). But after applying correction for multiple testing, statistical significance was retained only for HPV positive cases vs. HPV negative cases with Pc value of 0.006 (Pc for HPV positive cases vs. controls=0.07).
LTA +252 (rs909253) (AG/GG) genotype was also found to be more frequent in cases than controls with respect to carrier genotype (+252 AG/GG) distribution for total cases vs. controls (P=0.001), cancer vs. controls (P=0.003) and precancer vs. controls (P=0.042). However, after applying Bonferroni’s correction, the significance was retained for total cases vs. controls (Pc =0.006) and cancer vs. controls (Pc =0.018). +252 (A/G) LTA genotype with respect to HPV status showed an increased risk for HPV infection in women carrying the polymorphic allele (P = 0.004) for HPV positive cases vs. controls and for HPV negative vs. controls (P = 0.027). But after applying correction for multiple testing, significance was retained only for HPV positive vs. controls (Pc =0.024). The frequency distribution for +252 GG genotype (polymorphic homozygous) revealed highly significant association for HPV negative cases when compared to either controls or HPV positive cases with Pc value of 0.002 in the former and <0.001 in the latter. No association was found for TNFA -238 G/A (rs361525) and TNFA -863 C/A (rs1800630) polymorphisms either with respect to disease severity or HPV infection status.
Linkage Disequilibrium
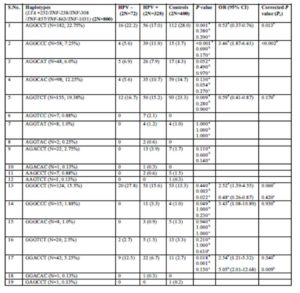
Combined analysis of both patient and control groups revealed that complete linkage disequilibrium was established between TNF -308 G/A and TNF -857 C/T (D’ = 1.0, r2 = 2) (Figure 1). Haplotype analysis using statistical software, PHASE, showed the presence of 19 haplotypes in 200 cervical cases including precancer and cancer and 200 controls (Table 2; Supplementary Table 2). However, only 12 haplotypes were present at frequencies ≥1% in the study population. Four haplotypes– AGGCCT (22.75%; 182/800), AGGCAC (12.25%; 98/800), AGGTCT (19.38%; 155/800), GGGCCT (15.5%; 124/800) were the most frequent.
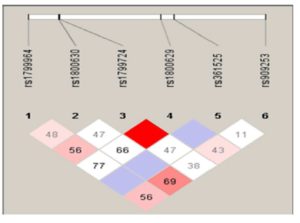
Value in each cell is the percent D’ between SNP pairs.
(rs1799724, rs1800629) — D’ 1.0; LOD 4.89; r2 0.029
TNF -LTA Haplotypes
The distribution of TNF-LTA haplotypes among cervical cancer, precancer and healthy controls (Table 2) revealed that three haplotypes were found to be significantly under-represented in case subjects in relation to controls–
AGGCCT (with wild alleles at all loci) in total cases vs. controls (P=0.001) and in cancer vs. controls (P=0.001), AGGCAC (with minor allele ‘A’ -863 locus) in total cases vs. controls (P=0.040) and in precancer vs. controls (P=0.015) and AGGTCT (with minor allele ‘T’ -857 locus) in total cases vs. controls (P=0.007) and in cancer vs. controls (P=0.006).
However, after the inclusion of correction for multiple testing, statistical significance was retained only for AGGCCT haplotype (with all wild alleles at all loci) for both total cases vs. controls (Pc =0.021) and cancer vs. controls (Pc =0.025), thereby establishing AGGCCT as the ‘protective haplotype.’
On the other hand, AGGCCC haplotype (with minor allele C for -1031T/C) was found to be significantly associated with total cases vs. controls (P=0.0002) and cancer vs. controls (P=0.0005) even after applying correction for multiple testing with Pc value of 0.004 in the former and 0.009 in the latter. Likewise, GGACCT (with minor alleles- A for -308 G/A & G for +252 A/G) was found to be significantly associated with total cases (Pc =0.042) and cancer (Pc =0.01) in comparison to controls. Hence, AGGCCC and GGACCT haplotypes can be considered to be important ‘risk/susceptibility haplotypes’ specifically for cervical cancer in the Indian population.
After stratifying the cases by HPV status (Supplementary Table 2), AGGCCT and AGGTCT showed a protective association with respect to HPV infection with a P value of 0.0007 in the former and 0.009 in the latter. But, statistical significance was retained only for AGGCCT (Pc=0.013 for AGGCCT and Pc=0.17 for AGGTCT) after applying the correction factor. On the other hand, AGGCCC was significantly associated with HPV positive cases in comparison to controls (P < 0.0001) even after applying correction for multiple testing (Pc <0.02).
Association of Plasma TNF and LTA Levels with Cervical Cancer/Precancer Cases
Plasma levels for both pro-inflammatory cytokines TNF and LTA were found to be significantly elevated in total cases (cervical cancer & precancer) in comparison to controls with a P value of < 0.001 for the former and 0.02 for the latter (Figure 2A & 2B). These levels were also found to be significantly associated with both precancer (P =0.045) and cancer subjects (P < 0.001) for TNF. But, interestingly, LTA levels were in strong association with precancerous lesions either with respect to controls (P = 0.001) or cancer cases (P = 0.01). TNF levels did not differ markedly between precancer and cancer.
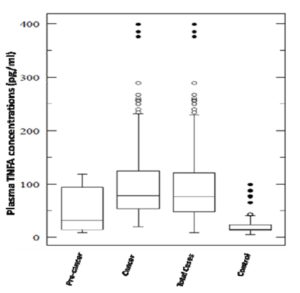
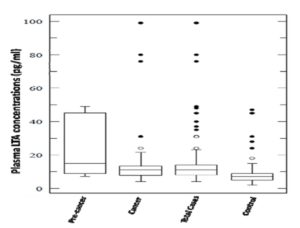
Association of Plasma TNF and LTA Levels with HPV Infection
The evaluation of the data according to HPV status revealed significantly higher plasma levels for both TNF and LTA in both the HPV positive (P < 0.0001) and HPV negative cases (P < 0.0001) than controls. An increasing trend from controls to HPV negative cases and then to HPV positive cases was obtained for both TNF and LTA, though the association was statistically insignificant (P= 0.97 for TNF; P= 0.10 for LTA in HPV+ cases vs. HPV- cases). Thus, plasma levels of these proinflammatory cytokines are independent of viral infections(data not shown).
Correlation of TNF/ LTA Polymorphisms with Plasma Levels:
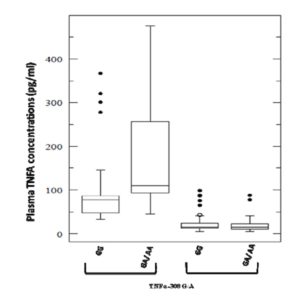
A functional correlation was established only for TNF -308 G/A polymorphism (rs1800629) with elevated plasma TNF levels in cases (P= 0.03 for GG vs. GA/AA), however the same could not be obtained in controls (Figure 3). No significant association with respect to genotype-levels was observed for the other polymorphisms or haplotypes (data not shown) in TNF-LTA locus.
Discussion
In India, cervical cancer is a leading cause of cancer deaths, contributing approximately one fourth of the total cancer incidence worldwide (Das et al. 2008). Approximately 10% of the healthy women are found to harbour HPV infection (Das et al. 2008), and among them 10%-30% women cannot clear HPV infection spontaneously, thus establishing the crucial role played by other factors including host-genetic, immunological and viral factors in HPV associated cervical carcinogenesis (Wang and Hildesheim 2003).
This genetic susceptibility can be attributed to the single nucleotide polymorphisms (SNPs) in the immunomodulatory TNF-LTA locus. TNF and LTA polymorphisms are reported to be associated with a wide range of inflammatory and immunomodulatory diseases including cancer (Wilson et al. 1997; Kaluza et al. 2000; Gonzalez et al. 2003; Ozaki et al. 2002; Deshpande et al. 2005). Much evidence supported that TNF/LTA polymorphisms were involved in the modulation of gene expression that affected the carcinogenesis (Wilson et al. 1997; Kaluza et al. 2000; Gonzalez et al. 2003; Ozaki et al. 2002; Deshpande et al. 2005; Hagihara et al. 1995). Variability in host immunogenetic background in the form of inherited polymorphisms in immunomodulatory genes may be an important determinant for conferring cell mediated immunity (CMI) to HPV mediated cervical cancer.
Therefore, in the current study we have evaluated the role of SNPs in TNF-LTA locus and their functional implications in cervical cancer among North Indian women. Although the current study is an extension of previous study (Kohaar et al. 2007), we report for the first time the association of TNF haplotypes and genotypes with HPV mediated cervical cancer and its prognosis in the Indian population.
Data on polymorphisms in TNF-LTA locus revealed that significant association was established, individually, for TNFA -308 G/A (rs1800629) and LTA +252 A/G (rs909253) SNPs with respect to total cases and invasive cervical cancer cases in comparison to controls. Therefore, TNFA -308 A and LTA +252 G alleles can be labelled as major susceptibility alleles for the development of cervical cancer with 2.89 and 1.6 folds increased risk among Indian women respectively.
Additionally, TNFA -308 G/A (rs1800629) and LTA +252 A/G (rs909253) polymorphisms were found to be an increased risk with respect to HPV infection. Interestingly, TNFA -1031 T/C SNP (rs1799964) was specifically related with HPV positive cervical cases in comparison to HPV negative cases. Conversely, TNFA -857 C/T SNP (rs1799724) was found to be associated with 54% reduced risk to the development of cervical cancer, thereby exhibiting a protective effect.
To further strengthen our findings on the role of SNPs in TNF-LTA locus, six locus haplotypes were constructed and their distribution was compared with respect to disease severity as well as HPV infection status. AGGCCT haplotype showed a protective association with respect to total cases and invasive cervical cancer group in comparison to controls. Similar protective association was established for AGGCCT haplotype in relation to HPV positive cervical cases. Hence, it can be inferred that AGGCCT haplotype might play an important role in enhancing the resolution of primary HPV infection or of cervical precancerous lesions. However, AGGCCC and GGACCT haplotypes emerged as important ‘risk/susceptibility haplotypes’ for total cervical cases and invasive cancer cases in comparison to controls with approximately 3 folds increased risk for both. On the same line, AGGCCC haplotype showed positive association for HPV infection in the Indian population; however GGACCT haplotype revealed significant association with HPV negative cases than controls.
Our study is in concordance with the studies on the Portuguese population (Duarte et al. 2005) in relation to -308 G/A polymorphism (rs1800629) and that on Korean population which revealed an increase of -308 A allele in cancer cases, although it was statistically insignificant (Jang et al. 2001). However, no significant difference was observed for -308 G/A polymorphism on the South African population (Black and Mixed race ethnic groups) (Govan et al. 2006), the U.S. population, (Calhoun et al. 2002) the Zimbabwean population (Stanczuk et al. 2003), and the Swedish population (Ghaderi et al. 2001). On the contrary, a report from the UK population showed that -308 G/A SNP was significantly under-represented in CIN patients (Kirkpatrick et al. 2004).
Similar to our previous findings, no significant association was observed for -238 G/A polymorphism (rs361525) and cervical cancer risk, although the frequency of heterozygotes was higher in controls, which is in line with a study on the Hispanic race of the US population (Wang and Hildesheim 2003). However, -238 SNP (rs361525) seemed to have a protective effect with respect to cervical cancer in the Korean (Jang et al. 2001) and US population (Calhoun et al. 2002).
Contradictory to our findings for -308 locus and in consensus with our results for -238 locus, no correlation was found for -238 G/A (rs361525) and -308 G/A (rs1800629) SNPs in relation to HPV status in the UK35 and US populations (Deshpande et al. 2005).
In line with our results, -857 C/T SNP (rs1799724) was also found to be significantly under-represented in HPV 16 positive cervical cases than controls in the Hispanic ethnic group (US). However, unlike our findings, -863 C/A SNP (rs1800630) also exhibited significant protective association in cases than controls in the Hispanic population (US) (Deshpande et al. 2005).
No prior studies are available to account for -1031T/C SNP (rs1799964) in TNFA promoter region and +252 A/G SNP (rs909253) in LTA gene with respect to cervical cancer. Ours is the only study in the context of the Indian population which extensively analyzed the SNPs in TNF-LTA locus, individually as well as in haplotypic association, in HPV associated cervical cancer. No other study except one by Deshpande et al on the Hispanic population (US) (Deshpande et al. 2005) has taken into consideration such an extensive mapping of TNF locus in relation to HPV infection and cervical cancer.
The epithelial cells of the cervix have been implicated in mucosal immune responses, for example by their ability to produce and secrete cytokines (Woodworth and Simpson 1993). Both TNF and LTA have been reported to exhibit cytostatic and cytotoxic effects on certain tumors (Aggarwal 2003; Mocellin and Nitti 2008; Vassalli 1992). The levels of the cytokines in circulation could affect the disease susceptibility and risk to HPV infection. TNFA and LTA bind to the same receptors i.e. TNF-R1 and R-2 receptors and share many biological activities (Pfeffer 2003). Additionally, at the genetic level, different LTA gene haplotypes have been associated with variations in the in vitro levels of TNFA production by inflammatory cells, probably due to extended linkage with the haplotypes in the TNFA gene (Koss et al. 2000). Thus, LTA indirectly mediates its effect by influencing TNFA levels.
In our study, higher circulating levels of both TNF and LTA are found in total cases vs. controls including cancer and precancer subjects. The plasma levels were also found to be significantly associated with precancer for LTA in comparison to controls. Regardless of HPV infection status, both TNF and LTA plasma levels were increased in cases than controls, which is in line with a previous study where no association was found between TNFA and HPV 16 (Ahmed et al. 2001). However, HPV 18 has been found to be associated with decreased intralesional TNFA levels (Fernandes et al. 2005). Our results are inconsistent with increased in-situ expression (Nicol et al. 2006) and elevated levels of TNFA in cervico-vaginal washings (Ahmed et al. 2001) and genital secretions (Passmore et al. 2007) of HPV mediated cancer of uterine cervix. On the same line, Chopra et al 1998, too, established an association for elevated circulating serum levels of both TNF and LTA cytokines with the progression of cervical cancer (Chopra et al. 1998). This is in agreement with a variety of studies, where plasma levels of TNFA or LTA have been shown to be associated with the outcome of cervical cancer and certain solid and haematological malignancies (Jang et al. 2001; Calhoun et al. 2002; Chopra et al. 1998; Song et al. 2007). On the other hand, our results are inconsistent with the study by Fernandes et al (Fernandes et al. 2005) and Song et al, 2007 (Song et al. 2007) with respect to TNF-α levels and cervical cancer.
Modifications of TNFA expression by variation in the TNFA promoter might interfere with viral clearance or promotes viral persistence that is a key factor underlying the phenomenon of cervical carcinogenesis. It is also noteworthy that a functional correlation was established between TNF -308 G/A polymorphism (rs1800629) and elevated plasma TNF levels in cases (P= 0.03 for GG vs. GA/AA). This is supportive of prior studies relating -308 A allele with higher inducible levels of gene transcription (Kroeger et al. 1997; Wilson et al. 1997) and TNFA protein production (Stuber et al. 1996; Davies et al. 2000). This SNP is located within a consensus sequence of transcription factor AP-2 and -308 A allele of the SNP has been associated with higher degree of transcription (Wilson et al. 1997). However, few studies showed no change either on TNF transcription or production (Stuber et al. 1996; Brinkman et al. 1995). However, no genotype—level association was established for other SNPs in TNFA promoter (rs361525, rs1799724, rs1800630, rs1799964) and rs909253 SNP in LTA.
Also, LTA +252 A/G SNP (rs909253) was found to be associated with increased TNFA secretary capacity as well as higher circulating levels of TNFA (Stuber et al. 1996). Even at the level of haplotypes, no association was achieved with respect to TNF-LTA levels, however few previous studies revealed positive association for the same (Davies et al. 2000).
Different results are obtained in different populations with different genetic backgrounds. The study may help in the future to understand the role of ethnic and geographical factors in disease pathogenesis as variable results are observed in different populations.
In summary, the present study revealed the potential role of genetic variations in immuno-modulatory TNF-LTA locus with HPV associated cervical cancer in the Indian population. Here, we report risk association for TNFA -308 (rs1800629) and LTA +252 SNP (rs909253) and the protective effect for TNFA -857 polymorphism (rs1799724) with the susceptibility to cervical cancer and HPV infection. Haplotype analysis revealed AGGCCC and GGACCT haplotypes as important ‘risk/susceptibility haplotypes’ and AGGCCT haplotype as a ‘protective haplotype’ for cervical cancer risk. Plasma levels were found to be elevated in both precancer and cancer subjects for TNF, while they were significantly associated with precancer for LTA. Genotype-level association revealed a functional correlation between TNF -308 G/A polymorphism (rs1800629) and elevated plasma TNF levels in the cases. However, in future replication studies, independent studies from various ethnic groups and further functional analysis are required to validate our findings. Inclusion of cytological normal but HPV positive individuals as controls will also strengthen the study.
Acknowledgements
The authors thank patients, their relatives and clinicians for their support and cooperation. The authors are also grateful to Council of Scientific and Industrial Research (CSIR) [60(0075)/06/EMR-II], Govt. of India for financial support.
References
1. Aggarwal, B. B. (2003). “Signalling pathways of the TNF superfamily: a double-edged sword.” Nat. Rev. Immunol., 3(9), 745-756.
Publisher – Google Scholar
2. Ahmed, M. I., Salahy, E. E., Fayed, S. T., El-Hefnawy, N. G., and Khalifa, A. (2001). “Human papillomavirus infection among Egyptian females with cervical carcinoma: relationship to spontaneous apoptosis and TNF-alpha.” Clin. Biochem., 34(6), 491-498.
Publisher – Google Scholar
3. Balkwill, F. (2002). “Tumor necrosis factor or tumor promoting factor?” Cytokine Growth Factor Rev., 13(2), 135-141.
Google Scholar
4. Bequet-Romero, M., and Lopez-Ocejo, O. (2000). “Angiogenesis modulators expression in culture cell lines positives for HPV-16 oncoproteins.” Biochem. Biophys. Res. Commun., 277(1), 55-61.
Publisher – Google Scholar
5. Beutler, B., and Bazzoni, F. (1998). “TNF, apoptosis and autoimmunity: a common thread?” Blood Cells Mol. Dis., 24(2), 216-230.
Publisher – Google Scholar
6. Brinkman, B. M., Zuijdeest, D., Kaijzel, E. L., Breedveld, F. C., and Verweij, C. L. (1995). “Relevance of the tumor necrosis factor alpha (TNF alpha) -308 promoter polymorphism in TNF alpha gene regulation.” J. Inflamm., 46(1), 32-41.
7. Calhoun, E. S., McGovern, R. M., Janney, C. A., Cerhan, J. R., Iturria, S. J., Smith, D. I., Gostout, B. S., and Persing, D. H. (2002). “Host genetic polymorphism analysis in cervical cancer.” Clin. Chem., 48(8), 1218-1224.
Google Scholar
8. Chopra, V., Dinh, T. V., and Hannigan, E. V. (1998). “Circulating serum levels of cytokines and angiogenic factors in patients with cervical cancer.” Cancer Invest, 16(3), 152-159.
Publisher – Google Scholar
9. Das, B. C., Hussain, S., Nasare, V., and Bharadwaj, M. (2008). “Prospects and prejudices of human papillomavirus vaccines in India.” Vaccine, 26(22), 2669-2679.
Publisher – Google Scholar
10. Das, B. C., Sharma, J. K., Gopalkrishna, V., Das, D. K., Singh, V., Gissmann, L., zur, H. H., and Luthra, U. K. (1992). “A high frequency of human papillomavirus DNA sequences in cervical carcinomas of Indian women as revealed by Southern blot hybridization and polymerase chain reaction.” J. Med. Virol., 36(4), 239-245.
Publisher – Google Scholar
11. Davies, F. E., Rollinson, S. J., Rawstron, A. C., Roman, E., Richards, S., Drayson, M., Child, J. A., and Morgan, G. J. (2000). “High-producer haplotypes of tumor necrosis factor alpha and lymphotoxin alpha are associated with an increased risk of myeloma and have an improved progression-free survival after treatment.” J. Clin. Oncol., 18(15), 2843-2851.
Google Scholar
12. Deshpande, A., Nolan, J. P., White, P. S., Valdez, Y. E., Hunt, W. C., Peyton, C. L., and Wheeler, C. M. (2005).“TNF-alpha promoter polymorphisms and susceptibility to human papillomavirus 16-associated cervical cancer.” J. Infect. Dis., 191(6), 969-976.
Publisher – Google Scholar
13. Duarte, I., Santos, A., Sousa, H., Catarino, R., Pinto, D., Matos, A., Pereira, D., Moutinho, J., Canedo, P., Machado, J. C., and Medeiros, R. (2005). “G-308A TNF-alpha polymorphism is associated with an increased risk of invasive cervical cancer.” Biochem. Biophys. Res. Commun., 334(2), 588-592.
Publisher – Google Scholar
14. Fernandes, A. P., Goncalves, M. A., Duarte, G., Cunha, F. Q., Simoes, R. T., and Donadi, E. A. (2005). “HPV16, HPV18, and HIV infection may influence cervical cytokine intralesional levels.” Virology, 334(2), 294-298.
Publisher – Google Scholar
15. Ghaderi, M., Nikitina, Z. L., Wallin, K., Wiklund, F., Hallmans, G., Lenner, P., Dillner, J., and Sanjeevi, C. B. (2001).“Tumor necrosis factor A and MHC class I chain related gene A (MIC-A) polymorphisms in Swedish patients with cervical cancer.” Hum. Immunol., 62(10), 1153-1158.
Publisher – Google Scholar
16. Gonzalez, S., Rodrigo, L., Martinez-Borra, J., Lopez-Vazquez, A., Fuentes, D., Nino, P., Cadahia, V., Saro, C., Dieguez, M. A., and Lopez-Larrea, C. (2003). “TNF-alpha -308A promoter polymorphism is associated with enhanced TNF-alpha production and inflammatory activity in Crohn’s patients with fistulizing disease.” Am. J. Gastroenterol., 98(5), 1101-1106.
Publisher – Google Scholar
17. Govan, V. A., Constant, D., Hoffman, M., and Williamson, A. L. (2006). “The allelic distribution of -308 Tumor Necrosis Factor-alpha gene polymorphism in South African women with cervical cancer and control women.” BMC. Cancer, 6, 24.
Publisher – Google Scholar
18. Hagihara, M., Shimura, T., Sato, K., Genga, K., Suzuki, M., and Tsuji, K. (1995). “HLA and tumor necrosis factor beta gene polymorphisms in Okinawa lung cancer patients: comparative study with mainland Japan lung cancer patients.” Hum. Immunol., 43(2), 95-100.
Publisher – Google Scholar
19. Jang, W. H., Yang, Y. I., Yea, S. S., Lee, Y. J., Chun, J. H., Kim, H. I., Kim, M. S., and Paik, K. H. (2001). “The -238 tumor necrosis factor-alpha promoter polymorphism is associated with decreased susceptibility to cancers.” Cancer Lett., 166(1), 41-46.
Publisher – Google Scholar
20. Kaluza, W., Reuss, E., Grossmann, S., Hug, R., Schopf, R. E., Galle, P. R., Maerker-Hermann, E., and Hoehler, T. (2000). “Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphism.” J. Invest Dermatol., 114(6), 1180-1183.
Publisher – Google Scholar
21. Kirkpatrick, A., Bidwell, J., van den Brule, A. J., Meijer, C. J., Pawade, J., and Glew, S. (2004).
“TNFalpha polymorphism frequencies in HPV-associated cervical dysplasia.” Gynecol. Oncol., 92(2), 675-679.
Publisher – Google Scholar
22. Knight, B., Yeoh, G. C., Husk, K. L., Ly, T., Abraham, L. J., Yu, C., Rhim, J. A., and Fausto, N. (2000).
“Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice.” J. Exp. Med., 192(12), 1809-1818.
Google Scholar
23. Kohaar, I., Thakur, N., Salhan, S., Batra, S., Singh, V., Sharma, A., Sodhani, P., Das, B. C., Sarkar, D. P., and Bharadwaj, M. (2007). “TNFalpha-308G/A polymorphism as a risk factor for HPV associated cervical cancer in Indian population.” Cell Oncol., 29(3), 249-256.
Google Scholar
24. Koss, K., Satsangi, J., Fanning, G. C., Welsh, K. I., and Jewell, D. P. (2000).
“Cytokine (TNF alpha, LT alpha and IL-10) polymorphisms in inflammatory bowel diseases and normal controls: differential effects on production and allele frequencies.” Genes Immun., 1(3), 185-190.
Publisher – Google Scholar
25. Kroeger, K. M., Carville, K. S., and Abraham, L. J. (1997). “The -308 tumor necrosis factor-alpha promoter polymorphism effects transcription.” Mol. Immunol., 34(5), 391-399.
Publisher – Google Scholar
26. Malejczyk, J., Malejczyk, M., Kock, A., Urbanski, A., Majewski, S., Hunzelmann, N., Jablonska, S., Orth, G., and Luger, T. A. (1992). “Autocrine growth limitation of human papillomavirus type 16-harboring keratinocytes by constitutively released tumor necrosis factor-alpha.” J. Immunol., 149(8), 2702-2708.
Google Scholar
27. Messer, G., Spengler, U., Jung, M. C., Honold, G., Blomer, K., Pape, G. R., Riethmuller, G., and Weiss, E. H. (1991). “Polymorphic structure of the tumor necrosis factor (TNF) locus: an NcoI polymorphism in the first intron of the human TNF-beta gene correlates with a variant amino acid in position 26 and a reduced level of TNF-beta production.”
J. Exp. Med., 173(1), 209-219.
Publisher – Google Scholar
28. Mocellin, S., and Nitti, D. (2008). “TNF and cancer: the two sides of the coin.” Front Biosci., 13, 2774-2783.
29. Moore, R. J., Owens, D. M., Stamp, G., Arnott, C., Burke, F., East, N., Holdsworth, H., Turner, L., Rollins, B., Pasparakis, M., Kollias, G., and Balkwill, F. (1999). “Mice deficient in tumor necrosis factor-alpha are resistant to skin carcinogenesis.” Nat. Med., 5(7), 828-831.
Google Scholar
30. Nicol, A. F., Nuovo, G. J., Wang, Y., Grinsztejn, B., Tristao, A., Russomano, F., Perez, M. A., Lapa e Silva JR, Fernandes, A. T., Gage, J. R., Martinez-Maza, O., and Bonecini-Almeida, M. G. (2006). “In situ detection of SOCS and cytokine expression in the uterine cervix from HIV/HPV coinfected women.” Exp. Mol. Pathol., 81(1), 42-47.
Publisher – Google Scholar
31. Ozaki, K., Ohnishi, Y., Iida, A., Sekine, A., Yamada, R., Tsunoda, T., Sato, H., Sato, H., Hori, M., Nakamura, Y., and Tanaka, T. (2002). “Functional SNPs in the lymphotoxin-alpha gene that are associated with susceptibility to myocardial infarction.” Nat. Genet., 32(4), 650-654.
Publisher – Google Scholar
32. Parkin, D. M., and Bray, F. (2006). “Chapter 2: The burden of HPV-related cancers.” Vaccine, 24 Suppl 3, S3-11-S3/25.
Google Scholar
33. Passmore, J. A., Morroni, C., Shapiro, S., Williamson, A. L., and Hoffman, M. (2007). “Papanicolaou smears and cervical inflammatory cytokine responses.” J. Inflamm. (Lond), 4, 8.
Google Scholar
34. Pfeffer, K. (2003). “Biological functions of tumor necrosis factor cytokines and their receptors.” Cytokine Growth Factor Rev., 14(3-4), 185-191.
Publisher – Google Scholar
35. Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B., and Erlich, H. A. (1988). “Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase.” Science, 239(4839), 487-491.
Google Scholar
36. Saiki, R. K., Scharf, S., Faloona, F., Mullis, K. B., Horn, G. T., Erlich, H. A., and Arnheim, N. (1985). “Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia.” Science, 230(4732), 1350-1354.
Google Scholar
37. Sambrook, J., Fritish, E. F., and Maniatis, T. Molecular Cloning: A Laboratory Manual. 9.16-9.19. 1989. Cold Spring Harbor Laboratory Press, Cold Spring Harbor NY. Ref Type: Generic
38. Sasieni, P. D. (1997). “From genotypes to genes: doubling the sample size.” Biometrics, 53(4), 1253-1261.
Publisher – Google Scholar
39. Skoog, T., van’t Hooft, F. M., Kallin, B., Jovinge, S., Boquist, S., Nilsson, J., Eriksson, P., and Hamsten, A. (1999). “A common functional polymorphism (C–>A substitution at position -863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha.” Hum. Mol. Genet., 8(8), 1443-1449.
40. Song, S. H., Lee, J. K., Seok, O. S., and Saw, H. S. (2007). “The relationship between cytokines and HPV-16, HPV-16 E6, E7, and high-risk HPV viral load in the uterine cervix.” Gynecol. Oncol., 104(3), 732-738.
Publisher – Google Scholar
41. Stanczuk, G. A., Sibanda, E. N., Tswana, S. A., and Bergstrom, S. (2003). “Polymorphism at the -308-promoter position of the tumor necrosis factor-alpha (TNF-alpha) gene and cervical cancer.” Int. J. Gynecol. Cancer, 13(2), 148-153.
Publisher – Google Scholar
42. Stuber, F., Petersen, M., Bokelmann, F., and Schade, U. (1996). “A genomic polymorphism within the tumor necrosis factor locus influences plasma tumor necrosis factor-alpha concentrations and outcome of patients with severe sepsis.” Crit Care Med., 24(3), 381-384.
Google Scholar
43. Tjiong, M. Y., van, d., V, ter Schegget, J. S., Burger, M. P., ten Kate, F. W., and Out, T. A. (2001). “Cytokines in cervicovaginal washing fluid from patients with cervical neoplasia.” Cytokine, 14(6), 357-360.
Publisher – Google Scholar
44. Vassalli, P. (1992). “The pathophysiology of tumor necrosis factors.” Annu. Rev. Immunol., 10, 411-452.
Publisher – Google Scholar
45. Wang, S. S., and Hildesheim, A. (2003). “Chapter 5: Viral and host factors in human papillomavirus persistence and progression.” J. Natl. Cancer Inst. Monogr,(31), 35-40.
Publisher – Google Scholar
46. Wilson, A. G., Clay, F. E., Crane, A. M., Cork, M. J., and Duff, G. W. (1995). “Comparative genetic association of human leukocyte antigen class II and tumor necrosis factor-alpha with dermatitis herpetiformis.” J. Invest Dermatol., 104(5), 856-858.
Publisher – Google Scholar
47. Wilson, A. G., Symons, J. A., McDowell, T. L., McDevitt, H. O., and Duff, G. W. (1997). “Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation.” Proc. Natl. Acad. Sci. U. S. A, 94(7), 3195-3199.
Publisher – Google Scholar
48. Woodworth, C. D., and Simpson, S. (1993). “Comparative lymphokine secretion by cultured normal human cervical keratinocytes, papillomavirus-immortalized, and carcinoma cell lines.” Am. J. Pathol., 142(5), 1544-1555.
Google Scholar
49. zur, H. H. (2002). “Papillomaviruses and cancer: from basic studies to clinical application.” Nat. Rev. Cancer, 2(5), 342-350.
Publisher – Google Scholar
Table 1: Distribution frequencies of SNPs in TNF-LTA locus among cervical cancer, precancer and healthy control subjects
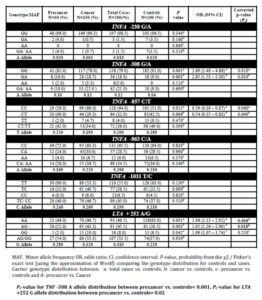
Table 2: Distribution frequencies of TNF-LTA haplotypes among cervical cancer, precancer and healthy controls
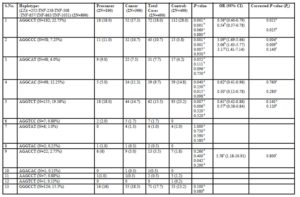
OR, odds ratio; CI, confidence interval; P-value, probability from the χ2 / Fisher’s exact test (using the approximation of Woolf) comparing the haplotype distribution for controls and cases. Haplotype distribution between: a- total cases vs. controls; b- cancer vs. controls, c- precancer vs. controls and d- precancer vs. cancer
Supplementary Table 1: Distribution frequencies of single nucleotide polymorphisms in TNF-LTA locus among HPV positive and HPV negative cervical cases and healthy control subjects
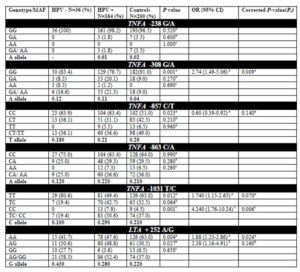
MAF, Minor allele frequency, OR, odds ratio; CI, confidence interval; P-value, probability from the χ2/ Fisher’s exact test (using the approximation of Woolf) comparing the genotype distribution for controls and cases. Carrier genotype distribution between: a- HPV positive cases vs. controls; b- HPV negative cases vs. controls and c- HPV positive cases vs. HPV negative cases. Pc-value for TNF -308 A allele distribution between HPV positive cases vs. controls= 0.01, Pc-value for TNF -1031 C allele distribution between HPV positive cases vs. HPV negative cases = 0.01, Pc-value for LTA +252 G allele distribution between HPV negative cases vs. controls = 0.001
Supplementary Table 2: Distribution frequencies of TNF-LTA haplotypes among HPV positive and HPV negative cervical cases (precancer and cancer) and healthy control subjects
OR, odds ratio; CI, confidence interval; P-value, probability from the χ2/ Fisher’s exact test (using the approximation of Woolf) comparing the haplotype distribution for controls and cases. Haplotype distribution between: a- HPV positive cases vs. controls; b- HPV negative cases vs. controls and c- HPV positive cases vs. HPV negative cases.