The isolation rate for E. coli (40%) in the current study is somewhat similar to that reported in captive green iguanas in Trinidad and Tobago where 60% captive green iguanas tested positive for E. coli. That same study showed that 58% of free-ranging mammals yielded E. coli and 90% of free-flying birds were positive for E. coli (Adesiyun, 1999). In a study conducted in Australia on non-mammalian vertebrates, 33% of crocodiles, 4% of turtles, 2% of snakes and 10% of lizards tested positive for E. coli (Gordon and Cowling, 2003). E. coli is documented to cause health problems in wildlife species, but this is considered rare (Adesiyun, 1999). E. coli has been reported as a cause of diarrhea in a tortoise (Owuamanam et al. 2012). Free-flying birds and free-ranging mammals harbor more E. coli in their gastrointestinal tract than reptiles do (Adesiyun, 1999).
Furthermore, 29% of the E. coli isolates in the present study were negative for sorbitol fermentation on sorbitol-MacConkey agar. E. coli O157:H7 is typically a non-sorbitol fermenter, however other zoonotic pathogenic non-O157 strains of E. coli are also non-sorbitol fermenters. Some pathogenic non-O157 strains of E. coli including O26, O103 and O111, have been previously associated with infections in humans. There is a possibility that there are other pathogenic Shiga-toxin producing E. coli strains among the isolates, however this study focused on E. coli O157:H7 since it is of paramount public health importance (Adesiyun, 1999). Additionally, the most commonly isolated E. coliserotype implicated in hemorrhagic colitis disease outbreaks in humans globally is E. coli O157:H7 and it serves as a marker for virulent strains of E. coli in various species (Nataro and Kaper, 1998, Perna et al., 2001).
E. coli O157:H7 was not identified in this study using Latex Agglutination Test Kits. In Trinidad and Tobago, all samples from 5 green iguanas fermented sorbitol and also tested negative for E. coli O157:H7 on Latex Agglutination Test. Free-ranging and captive avian, reptile and mammalian wildlife species were not important reservoirs of E. coliO157:H7 in Trinidad and Tobago (Adesiyun, 1999). Similarly, it appears that green iguanas in Grenada do not harborE. coli O157:H7 and may therefore not pose a public health in this regard. However, it may be noted that other methods of detecting E. coli O157:H7, including PCR are available but they were not employed in this study.
Although there is no information on Shiga-toxin producing E. coli in green iguanas in England, wild rabbits appear to be significant reservoirs of E. coli O157:H7 in that country; 16.2% fecal samples from wild rabbits tested positive for Shiga-toxin producing E. coli and 6.2% of said fecal samples were confirmed as E. coli O157:H7 (Sciafe and Crook, 2005). In a study in the United States, using culture, latex agglutination and PCR, 521 fecal samples from wild life species including raccoons (230 samples), deer (141 samples), opossums (25 samples), birds (9 samples), and other species (16 samples) were analyzed; only one (0.19%) wild opossum of the 521 samples tested positive for E. coli O157 (Renter et al., 2003). E. coli O157:H7 was isolated from 23.4% feral pigs in California, USA. This study confirmed that mammalian wild life species in California, USA may be implicated in cross contamination of vegetables destined for human consumption and therefore constitute an important public health source of pathogenic E. coli (Jay-Russell, 2010). In another study conducted in three counties in California, USA, 730 samples from various wild life species including raccoons, opossums, striped skunk and tule elk tested negative for E. coliO157:H7 (Mandrell et al., 2010). Obviously, wildlife species other than rabbits and feral pigs are not common reservoirs of this serotype of Shiga-toxin producing E. coli (Renter et al., 2003).
Green iguanas have however been shown to be significant sources of zoonotic pathogenic Salmonella in Grenada (Sylvester et al., 2013). It is possible that in the current study the prevalence of E. coli O157:H7 is underestimated since PCR and/or serotyping of E. coli were not performed. Due to the frequent human-wildlife interactions, the proximity of human and wildlife habitats as well as the tendency for Grenadians to consume wild meat, there is an urgent need to conduct continued research in wildlife species in Grenada. Future studies should include use of PCR and serotyping.
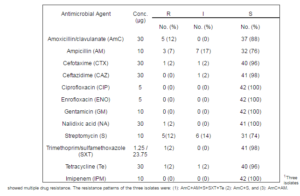
In this current study antibiotic resistance was detected for seven of the twelve antibiotics used. The highest resistance was 12%, recorded for amoxicillin/clavalunic acid and streptomycin, followed by ampicillin and nalidixic acid, cefotaxime, tetracycline and trimethoprim/sulfamethoxazole. It is interesting to note that resistance was found in multiple antimicrobial drug categories/classes including penicillins, tetracyclines, cephalosporins, aminoglycosides, sulfanomide-trimethoprim and quinolones (Table 2). Additionally, it should be considered significant that one E. coliisolate showed intermediate resistance to two third generation cephalosporins (cefotaxime and ceftazidime) used in treating complicated systemic bacterial infections (Pournaras et al., 2004). This antibiotic resistance pattern differs to that reported in Salmonella isolates from green iguanas, blue land crabs and cane toads in Grenada where no resistance was reported (Sylvester et al., 2013; Peterson et al., 2013; Drake et al., 2012). In a study of 54 E. coliisolates from feral cats in Grenada, no resistance was found in 65% of the isolates tested against 6 drugs. Resistance to amoxicillin-clavulanic acid was found only in 2% of the isolates (Hariharan et al., 2011). The relatively high antibiotic resistance amongst E. coli isolates from green iguanas is of public health concern since these resistance genes may be transferred to other non-pathogenic and/or pathogenic E. coli and other members ofEnterobacteriaceae, including Salmonella in the environment or in the human gut. We propose that future studies onE. coli in wildlife should include determination of minimal inhibitory concentrations of antimicrobial drugs for critical analysis of drug resistance.
Conclusions
In this study E. coli O157:H7 was not detected in green iguanas in Grenada. The antibiotic resistance patterns found in this study are of public health concern since there are ample opportunities for transmission of pathogenic and/or commensal microorganisms from iguanas to humans, particularly because green iguanas are used for human consumption in Grenada.
Acknowledgements
This project was reviewed and approved by The St George’s University Institutional Animal Care and Use Committee (IACUC-10011-R).
References
Adesiyun, A. A. (1999). “Absence of Escherichia coli O157 in a survey of Wildlife from Trinidad and Tobago,” Journal of Wildlife Diseases. 35, 115-120.
Publisher – Google Scholar
Alberts, A. C., Carter, R. L., Hayes, W. K. & Martin, E. P. (2004). Iguanas; Biology and Conservation, University of California Press, Berkeley and Los Angeles, CA.
Publisher – Google Scholar
Drake, M., Amadi, V., Zieger, U., Johnson, R. & Hariharan, H. (2012). “Prevalence of Salmonella spp. in Cane Toads (Bufo Marinus) from Grenada, West Indies, and Their Antimicrobial Susceptibility,” Zoonoses and Public Health. 60, 437-441.
Publisher – Google Scholar
Gordon, D. M. & Cowling, A. (2003). “The Distribution and Genetic Structure of Escherichia Coli in Australian Vertebrates: Host and Geographic Effects,” Microbiology. 149, 3575-3586.
Publisher – Google Scholar
Hariharan, H., Matthew, V., Fountain, J., Snell, A., Doherty, D., King, B., Shemer, E., Oliveira, S. & Sharma, R. N. (2011). “Aerobic Bacteria from Mucous Membranes, Ear Canals, and Skin Wounds of Feral Cats in Grenada, and the Antimicrobial Drug Susceptibility of Major Isolates,” Comparative Immunology, Microbiology and Infectious Diseases. 34, 129-134.
Publisher – Google Scholar
Jay-Russell, M. (2010). “E. Coli O157 in Central Coast Wildlife,” University of California Cooperative Extension. California, USA. Retrieved from: cesanluisobispo.ucanr.edu/files/163895.pdf
Publisher
Jorgensen, J. H. & Turnidge, J. D. (2003). ‘Susceptibility Test Methods: Dilution and Disk Diffusion Methods,’ In: P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, R. H. Yolken (Eds.), Manual of Clinical Microbiology, (8th ed), American Society for Microbiology Press, Washington, DC. Pp 1108-1127.
Google Scholar
Mandrell, R., Cooley, M., Atwill, R., Jay-Russell, M., Larsen, R., Orthmeyer, D. & Chandler, S. (2010). “Ecology and Epidemiology of Escherichia Coli O157:H7 in Fresh Produce Production Regions of Salinas, California/Central California Coast, 2008-2010,” Retrieved from: cesanluisobispo.ucanr.edu/files/163895.pdf
Publisher
Mermin, J., Hoar, B. & Angulo, F. J. (1997). “Iguanas and Salmonella Marina infection in children: A Reflection of the Increasing Incidence of Reptile-Associated Salmonellosis in the United States,” Pediatrics, 99, 399-402.
Publisher – Google Scholar
Nataro, J. P. & Kaper, J. B. (1998). “Diarrheagenic Escherichia Coli,” Clinical Microbiology Revews. 11, 142-201.
Publisher – Google Scholar
Owuamanam, C. M., Ameen S. A. & Adedokun, R. A. M. (2012). “Clinical Case Report on Diarrhea Caused by Escherichia Coli in Horsfields Tortoise (Testudo Horsfieldi) at Zoological Garden, University of Ibadan, Nigeria,” Global Veterinaria 9, 717-719.
Publisher
Perna, N. T., Plunkett, G.III., Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., Pósfai, G., Hackett, J., Klink, S., Boutin, A., Shao, Y., Miller, L., Grotbeck, E. J., Davis, N. W., Lim, A., Dimalanta, E. T., Potamousis, K. D., Apodaca, J., Anantharaman, T. S., Lin, J., Yen, G., Schwartz, D. C., Welch, R. A. & Blattner, F. R. (2001). “Genome Sequence of Enterohaemorrhagic Escherichia Coli O157:H7,” Nature 409, 529-533.
Publisher – Google Scholar
Peterson, R., Hariharan, H., Matthew, V., Chappell, S., Davies, R., Parker, R. & Sharma, R. (2013). “Prevalence, Serovars, and Antimicrobial Susceptibility of Salmonella Isolated from Blue Land Crabs (Cardisoma Guanhumi) in Grenada, West Indies,” Journal of Food Protection 76, 1270-1273.
Publisher – Google Scholar
Pournaras, S., Ikonomidis, A., Sofianou D., Tsakris, A. & Maniatis, A. N. (2004). “CTX-M-Type B-Lactamases Affect Community Escherichia Coli Treatment, Greece,” Emerging Infectious Diseases. Retrieved from:http://wwwnc.cdc.gov/eid/article/10/6/03-1031.htm
Publisher
Quinn, P. J., Markey, B. K., Leonard, F. C., FitzPatrick, E. S., Fanning, S. & Hartigan, P. A. (2011). Veterinary Microbiology and Microbial Disease. 2nd Edition, Wiley-Blackwell Publishing Company. Chichester, West Sussex, UK.
Publisher – Google Scholar
Renter, D. G., Sargeant, J. M., Oberst, R. D. & Samadpour, M. (2003). “Diversity, Frequency and Persistence of Escherichia Coli O157 Strains from Range Cattle Environments,” Applied and Environmental Microbiology. 69, 542-547.
Publisher – Google Scholar
Santos, A. C. M., Zidko, A. C. M., Pignatari, A. C. & Silva, R. M. (2013). “Assessing the Diversity of the Virulence Potential of Escherichia Coli Isolated from Bacteremia in Sao Paulo, Brazil,” Brazilian Journal of Medical and Biological Research. 46, 968-973.
Publisher – Google Scholar
Sciafe, H. & Crook, B. (2005). “Wild Rabbits as Potential Carriers of E. Coli VTEC-Final Report,” Health Sciences.Health & Safety Laboratory. Broad Lane, Sheffield, England, UK.
Publisher
Sylvester, W. R. B., Amadi, V., Pinckney, R., MacPherson, C., McKibben, J., Bruhl-Day, R., Johnson, R. & Hariharan, H. (2013). “Prevalence, Serovars, and Antimicrobial Susceptibility of Salmonella spp. from Wild and Domestic Green Iguanas (Iguana Iguana) from Grenada, West Indies,” Zoonoses and Public Health. Published online.
Publisher – Google Scholar
Taddei, S., Dodi, P. L., Di lanni, F., Cabassi, C. S. & Cavirani, S. (2010). “Conjunctival Flora of Clinically Normal Captive Green Iguanas (Iguana Iguana),” Veterinary Record.167, 29-30.
Publisher – Google Scholar