Introduction
Multiple sclerosis (MS) is a chronic demyelinating disorder of the central nervous system that results in neurodegeneration. No definitive triggers have been identified for this autoimmune disease; however, predisposition to MS is believed to be the result of a combination of various genetic and environmental factors (Disanto et al., 2012).
Several studies have associated MS with human leukocyte antigen (HLA) genes on the chromosome 6 p21.3 region. Associations between MS and HLA genotypes have been heterogeneous among the various populations tested; nevertheless, the most commonly reported association to date is with the HLA class II allele DRB1*1501 (Svejgaard, 2008). Genome-wide association studies have also revealed other genetic determinants that may play a role in susceptibility to MS. These include particular alleles of the interleukin-2 and interleukin-7 receptor α-chain genes, IL2RA and IL7RA respectively, among various others (Sawcer, 2008, Hafler et al., 2007, Oksenberg et al., 2008, Sawcer et al., 2011).
An association with low vitamin D levels has been prominent among the diverse environmental factors linked to MS (Berlanga-Taylor and Ramagopalan, 2013). Additionally, a myriad of potential infectious triggers have been investigated in relation to this disease with strong correlations reported between an Epstein-Barr virus (EBV) infection and MS (DeLorenze et al., 2006). The Human herpesvirus 6 (HHV-6) has also been reported as a risk factor for this disorder by various studies; however, credence of such a correlation was diminished by the irreproducibility of some these studies and by flaws in the study designs employed (Coates and Bell, 1998, Goldberg et al., 1999, Liedtke et al., 1995, Martin et al., 1997, Mirandola et al., 1999, Simpson et al., 2012, Gustafsson et al., 2013, Virtanen et al., 2014).
Our group has previously examined whether an HHV-6 infection increases the risk of MS (Fayad et al., 2014). HHV-6 belongs to the Herpesviridae, or the herpes family of viruses. These are DNA viruses that can cause latent and recurring infections. These viruses have various immune system-evasion mechanisms that allow them to establish life-long latency in the host. To assess whether a HHV-6 infection plays a role in the pathobiology of MS, blood samples from MS patients and controls were investigated for harboring HHV-6 DNA. We detected a correlation between this type of infection and MS (OR = 2.935, 95% CI = 1.582-5.463, p=0.000). Analysis of the rs2104286 and rs12722489 single nucleotide polymorphisms (SNPs) in the IL2RA gene and the rs6897932 SNP in the IL7RA gene did not reveal any statistically significant correlations with MS in our population; however, concomitant presence of the rs2104286 A-allele or the rs6897932 C-allele with HHV-6 moderately increased the risk of MS compared to HHV-6 alone.
In the study at hand, we set out to examine the effect of HLA genetic determinants on the risk of MS in complex with HHV-6 and particular SNPs in the IL2RA and IL7RA genes.
Materials and Methods
Study Population and Specimens
Blood samples were obtained from 41 MS patients (27 females and 14 males) and 38 controls (28 females and 10 males). All subjects were Lebanese, older than the age of 18 and were attending the Multiple Sclerosis Clinic and Research Center (Doctors Center, Beirut, Lebanon). The subject age range was 19-70 years with an average age of 39.76 ± 12.33. All patients fulfilled the Mc Donald’s criteria (McDonald et al., 2001) prior to enrollment. The control group included non-MS subjects reporting no autoimmune disorders in immediate family members or themselves. No subjects who were pregnant or on an immunosuppressive regimen were enrolled in the study. The Institutional Review Board (IRB) at the American University of Beirut (AUB) approved this study prior to its commencement and signed consent was obtained from all subjects enrolled. Blood samples were initially collected in citrated tubes and then DNA was obtained from blood utilizing phenol extraction and ethanol precipitation (Ausubel FM et al., 2000).
HLA Typing
HLA-A, B and C allele typing was performed utilizing the ABC SSP UniTray kit (Invitrogen, Carlsbad, CA) whereas HLA-DR and DQ allele typing was performed employing the DRDQ SSP UniTray kit (Invitrogen). Manufacturer procedure recommendations were followed.
HHV-6 and Interleukin Receptor Α-Chain Gene SNP Determination
HHV-6 positivity and detection of the IL2RA rs2104286 A-allele, IL7RA rs6897932 C- allele and the IL2RA rs12722489 G-allele were performed as previously described (Fayad et al., 2014).
Statistical Analysis
Statistical analysis was performed using PASW Statistics 18 for Windows.
Results
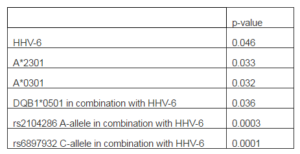
To explore the effect of HLA genetic determinants on susceptibility to MS, patients and matched controls were genotyped. Significant associations are described in Table 1. We detected a correlation between MS and the A*2301 allele (p=0.033) which was detected in 12.19% of patients but none of the controls (RR=2.056, 95% CI=0.913-2.056). We also observed a correlation between MS and the A*0301 allele (p=0.032) whereby 21.95% of the patients tested were positive for this allele compared to only 5.26% of the controls (RR=1.739, 95% CI=0.955-2.167). These associations were not secondary to linkage disequilibrium with other alleles since no such linkage was detected in our study population.
Including HHV-6 positivity into the analysis revealed that double positivity for the virus and the DQB1*0501 allele was associated with MS (p=0.036) whereby 17.07% of patients but only 2.63% of the controls were concomitantly positive for both factors (RR=1.827, 95%CI=0.926-2.134). This hence increased the risk exerted by DQB1*0501 alone (RR=1.495, 95%CI=0.903-2.138) which by itself was not statistically significant (p=0.06); this allele was detected in 36.58% of patients and 18.42% of controls. HHV-6 positivity by itself was significantly associated with MS (p=0.046) in the current study population whereby it was detected in 62.5% of patients and 39.47% of controls (RR=1.523, 95%CI=0.942-2.474).
On the other hand, a modest protective effect was observed for the DQB1*0302 allele (p=0.049) which was identified in 10.53% of controls but none of the patients.
Detection of the IL2RA rs2104286 A-allele, IL7RA rs6897932 C- allele and the IL2RA rs12722489 G-alleles double positivity with the various HLA genetic markers did not reveal any significant increases in the risk for MS. As we previously reported (Fayad et al., 2014), the IL2RA rs2104286 A-allele was present in 98% of MS patient and in 96% of control, the IL2RA rs12722489 G-allele was present in all subjects, whereas the IL7RA rs6897932 C-allele was present in 98% of MS patients and 93% of controls thus revealing no correlations with MS. Inclusion of HHV-6 positivity into the analysis did not yield any significant associations either. Hence, combinational positivity for these SNPs, HHV-6 and the various HLA alleles did not result in significant MS risk increases in our study population.
Table 1. Significant Associations with Multiple Sclerosis
Discussion
The DRB1*1501 allele is often indicated as the most common genetic determinant associated with MS. This association has been reported in African-American (McElroy et al., 2010), Turkish (SaruhanDireskeneli et al., 1997), Northern European (Fogdell et al., 1995) and other Caucasian populations (Oksenberg et al., 2008, McElroy and Oksenberg, 2011). In Caucasians, the association has been mapped to the DRB5*0101-DRB1*1501-DQA1*0102- DQB1*0602 haplotype since these alleles are in strong linkage disequilibrium in such populations. In the Lebanese population, as our data revealed, no such correlation exists. We rather detected as association with the A*2301 and A*0301 alleles instead. Other populations that have similarly observed no associations with the DRB1*1501 allele include African-Brazillians (Caballero et al., 1999) and Sardinians (Marrosu et al., 1998).
HLA-A3 (Naito et al., 1972) and HLA-B7 (Jersild et al., 1972) have been previously associated with MS in Caucasians, however it was later found that these associations were secondary to class II direct association with the disease and the result of HLA alleles being in linkage disequilibrium (Compston et al., 1976, Terasaki et al., 1976). As previously indicated the association between the described class I alleles and MS in our population are primary since no similar linkages were detected. Control HLA frequencies detected in this study were comparable to those previously reported in the Lebanese population (Abdelnoor et al., 2001).
On the other hand, we detect a modest protective effect for the DQB1*0302 on MS. Surprisingly, this allele has been reported to play a role in MS in the Sardinian population (Marrosu et al., 1992); however, DQB1*0302 is in linkage disequilibrium with other alleles in susceptibility-conferring haplotypes in Sardinians (Marrosu et al., 1998) which may explain why this allele was reported to be associated with MS in this particular population.
Inclusion of HHV-6 detection into our analysis revealed that double positivity for the virus and the DQB1*0501 allele enhanced the risk of MS. Whereas we had previously reported that HHV-6 infection by itself is a risk factor for MS (Fayad et al., 2014), DQB1*0501 positivity showed no such associations on its own in the Lebanese population. This allele does not enhance the risk of HHV-6 infection by itself as revealed by our analysis; thus, the increased MS risk is not secondary to any predisposition for infection imparted by the DQB1*0501 allele.
We also previously observed that particular SNPs in the IL2RA and IL7RA genes modestly enhance the risk of MS in subjects positive for HHV-6 (Fayad et al., 2014). However, our analysis for these SNPs in the current study did not reveal any susceptibilities for MS in conjunction with risk HLA genes and HHV-6. This may indicate that these interleukin receptor α-chain genes and HLA alleles are in an epistatic relation with each other or with another yet undetermined factor modulating their role in MS upon infection with HHV-6. The role played by HHV-6 in exacerbating or triggering MS has not been elucidated, nevertheless antigenic similarities between HHV-6 viral peptides and the myelin basic protein have been reported (Wucherpfennig and Strominger, 1995, Tejada-Simon et al., 2003). This may contribute in part to the autoimmune process resulting in MS.
The heterogeneity in disease associations with particular genetic make-ups remains population dependent as revealed by our study as well as by others. Therefore, any conclusions regarding risk should always take into account the ethnicity of the population tested. Our observations indicate that certain genetic determinants enhance the risk of MS in HHV-6 positive individuals. These genetic components do not result in the same effect in subjects not infected with this virus.
Acknowledgements
The authors wish to thank the Lebanese National Council for Scientific Research (CNRS) for funding this work.
References
Abdelnoor, A. M., Abdelnoor, M., Heneine, W., Khauli, R., Kobeissy, F., Mansur, S., Malak, R. & Sharara, H. (2001). “Major Histocompatibility Complex Class I and II Antigens Frequencies in Selected Groups of Lebanese,” Transplantation Proceedings, 33, 2839-40.
Publisher – Google Scholar
Ausubel, F. M., Brent, R, Kingston, R. E., Moore, D. D., S. J., Smith Ja & K, S. (Eds.) (2000). ‘Current Protocols in Molecular Biology,’ New York, NY: John Wiley and Sons, Inc.
Berlanga-Taylor, A. J. & Ramagopalan, S. V. (2013). “Vitamin D and Multiple Sclerosis: What is the Clinical Impact?,”Expert Opinion on Medical Diagnostics, 7, 227-9.
Publisher – Google Scholar
Caballero, A., Alves-Leon, S., Papais-Alvarenga, R., Fernandez, O., Navarro, G. & Alonso, A. (1999). “DQB1*0602 Confers Genetic Susceptibility to Multiple Sclerosis in Afro-Brazilians,” Tissue Antigens, 54, 524-6.
Publisher – Google Scholar
Coates, A. R. & Bell, J. (1998). “HHV-6 and Multiple Sclerosis,” Nature Medicine, 4, 537-8.
Publisher – Google Scholar
Compston, D. A., Batchelor, J. R. & McDonald, W. I. (1976). “B-Lymphocyte Alloantigens Associated with Multiple Sclerosis,” The Lancet, 2, 1261-5.
Publisher – Google Scholar
DeLorenze, G. N., Munger, K. L., Lennette, E. T., Orentreich, N., Vogelman, J. H. & Ascherio, A. (2006). “Epstein-Barr Virus and Multiple Sclerosis: Evidence of Association from a Prospective Study with Long-Term Follow-Up,” Archives of Neurology, 63, 839-44.
Publisher – Google Scholar
Disanto, G., Morahan, J. M. & Ramagopalan, S. V. (2012). “Multiple Sclerosis: Risk Factors and Their Interactions,”CNS & Neurological Disorders – Drug Targets, 11, 545-55.
Publisher – Google Scholar
Fogdell, A., Hillert, J., Sachs, C. & Olerup, O. (1995). “The Multiple Sclerosis- and Narcolepsy-Associated HLA Class II Haplotype Includes the DRB5*0101 Allele,” Tissue Antigens, 46, 333-6.
Publisher – Google Scholar
Goldberg, S. H., Albright, A. V., Lisak, R. P. & Gonzalez-Scarano, F. (1999). “Polymerase Chain Reaction Analysis of Human Herpesvirus-6 Sequences in the Sera and Cerebrospinal Fluid of Patients with Multiple Sclerosis,” Journal of Neurovirology, 5, 134-9.
Publisher – Google Scholar
Gustafsson, R., Reitsma, R., Stralfors, A., Lindholm, A., Press, R. & Fogdell-Hahn, A. (2013). “Incidence of Human Herpesvirus 6 in Clinical Samples from Swedish Patients with Demyelinating Diseases,” Journal of Microbiology, Immunology and Infection.
Publisher – Google Scholar
Hafler, D. A., Compston, A., Sawcer, S., Lander, E. S., Daly, M. J., De Jager, P. L., De Bakker, P. I., Gabriel, S. B., Mirel, D. B., Ivinson, A. J., Pericak-Vance, M. A., Gregory, S. G., Rioux, J. D., Mccauley, J. L., Haines, J. L., Barcellos, L. F., Cree, B., Oksenberg, J. R. & Hauser, S. L. (2007). “Risk Alleles for Multiple Sclerosis Identified by a Genomewide Study,” The New England Journal of Medicine, 357, 851-62.
Publisher – Google Scholar
Jersild, C., Svejgaard, A. & Fog, T. (1972). “HL-A Antigens and Multiple Sclerosis,” The Lancet, 1, 1240-1.
Publisher – Google Scholar
Liedtke, W., Malessa, R., Faustmann, P. M. & Eis-Hubinger, A. M. (1995). “Human Herpesvirus 6 Polymerase Chain Reaction Findings in Human Immunodeficiency Virus Associated Neurological Disease and Multiple Sclerosis,” Journal of Neurovirology, 1, 253-8.
Publisher – Google Scholar
Marrosu, M. G., Muntoni, F., Murru, M. R., Costa, G., Pischedda, M. P., Pirastu, M., Sotgiu, S., Rosati, G. & Cianchetti, C. (1992). “Hla-Dqb1 Genotype in Sardinian Multiple-Sclerosis – Evidence for a Key Role of Dqb1 Asterisk-0201 and Asterisk-0302 Alleles,” Neurology, 42, 883-886.
Publisher – Google Scholar
Marrosu, M. G., Murru, M. R., Costa, G., Murru, R., Muntoni, F. & Cucca, F. (1998). “DRB1-DQA1-DQB1 Loci and Multiple Sclerosis Predisposition in the Sardinian Population,” Human Molecular Genetics, 7, 1235-1237.
Publisher – Google Scholar
Martin, C., Enbom, M., Soderstrom, M., Fredrikson, S., Dahl, H., Lycke, J., Bergstrom, T. & Linde, A. (1997). “Absence of Seven Human Herpesviruses, Including HHV-6, by Polymerase Chain Reaction in CSF and Blood from Patients with Multiple Sclerosis and Optic Neuritis,” Acta Neurologica Scandinavica, 95, 280-3.
Publisher – Google Scholar
Mcdonald, W. I., Compston, A., Edan, G., Goodkin, D., Hartung, H. P., Lublin, F. D., Mcfarland, H. F., Paty, D. W., Polman, C. H., Reingold, S. C., Sandberg-Wollheim, M., Sibley, W., Thompson, A., Van Den Noort, S., Weinshenker, B. Y. & Wolinsky, J. S. (2001). “Recommended Diagnostic Criteria for Multiple Sclerosis: Guidelines from the International Panel on the Diagnosis of Multiple Sclerosis,” Annals of Neurology, 50, 121-7.
Publisher – Google Scholar
McElroy, J. P., Cree, B. A. C., Caillier, S. J., Gregersen, P. K., Herbert, J., Khan, O. A., Freudenberg, J., Lee, A., Bridges, S. L., Jr., Hauser, S. L., Oksenberg, J. R. & Gourraud, P. A. (2010). “Refining the Association of MHC with Multiple Sclerosis in African Americans,” Human Molecular Genetics, 19, 3080-8.
Publisher – Google Scholar
McElroy, J. P. & Oksenberg, J. R. (2011). “Multiple Sclerosis Genetics 2010,” Neurologic Clinics, 29, 219-31.
Publisher – Google Scholar
Mirandola, P., Stefan, A., Brambilla, E., Campadelli-Fiume, G. & Grimaldi, L. M. (1999). “Absence of Human Herpesvirus 6 and 7 from Spinal Fluid and Serum of Multiple Sclerosis Patients,” Neurology, 53, 1367-8.
Publisher – Google Scholar
Naito, S., Namerow, N., Mickey, M. R. & Terasaki, P. I. (1972). “Multiple Sclerosis: Association with HL-A3,” Tissue Antigens, 2, 1-4.
Publisher – Google Scholar
Oksenberg, J. R., Baranzini, S. E., Sawcer, S. & Hauser, S. L. (2008). “The Genetics of Multiple Sclerosis: SNPs to Pathways to Pathogenesis,” Nature Reviews Genetics, 9, 516-26.
Publisher – Google Scholar
Saruhandireskeneli, G., Esin, S., Baykan-kurt, B., Ornek, I., Vaughan, R. & Eraksoy, M. (1997). “HLA-DR and -DQ Associations with Multiple Sclerosis in Turkey,” Human Immunology, 55, 59-65.
Publisher – Google Scholar
Sawcer, S. (2008). “The Complex Genetics of Multiple Sclerosis: Pitfalls and Prospects,” Brain, 131, 3118-31.
Publisher – Google Scholar
Sawcer, S., Hellenthal, G., Pirinen, M., Spencer, C. C., Patsopoulos, N. A., Moutsianas, L., Dilthey, A., Su, Z., Freeman, C., Hunt, S. E., Edkins, S., Gray, E., Booth, D. R., Potter, S. C., Goris, A., Band, G., Oturai, A. B., Strange, A., Saarela, J., Bellenguez, C., Fontaine, B., Gillman, M., Hemmer, B., Gwilliam, R., Zipp, F., Jayakumar, A., Martin, R., Leslie, S., Hawkins, S., Giannoulatou, E., D’alfonso, S., Blackburn, H., Martinelli Boneschi, F., Liddle, J., Harbo, H. F., Perez, M. L., Spurkland, A., Waller, M. J., Mycko, M. P., Ricketts, M., Comabella, M., Hammond, N., Kockum, I., Mccann, O. T., Ban, M., Whittaker, P., Kemppinen, A., Weston, P., Hawkins, C., Widaa, S., Zajicek, J., Dronov, S., Robertson, N., Bumpstead, S. J., Barcellos, L. F., Ravindrarajah, R., Abraham, R., Alfredsson, L., Ardlie, K., Aubin, C., Baker, A., Baker, K., Baranzini, S. E., Bergamaschi, L., Bergamaschi, R., Bernstein, A., Berthele, A., Boggild, M., Bradfield, J. P., Brassat, D., Broadley, S. A., Buck, D., Butzkueven, H., Capra, R., Carroll, W. M., Cavalla, P., Celius, E. G., Cepok, S., Chiavacci, R., Clerget-Darpoux, F., Clysters, K., Comi, G., Cossburn, M., Cournu-Rebeix, I., Cox, M. B., Cozen, W., Cree, B. A., Cross, A. H., Cusi, D., Daly, M. J., Davis, E., De Bakker, P. I., Debouverie, M., D’hooghe M, B., Dixon, K., Dobosi, R., Dubois, B., Ellinghaus, D., Elovaara, I., Esposito, F. et al. (2011). “Genetic Risk and a Primary Role for Cell-Mediated Immune Mechanisms in Multiple Sclerosis,” Nature, 476, 214-9.
Publisher – Google Scholar
Simpson, S., Jr., Taylor, B., Dwyer, D. E., Taylor, J., Blizzard, L., Ponsonby, A. L., Pittas, F., Dwyer, T. & Van Der Mei, I. (2012). “Anti-HHV-6 Igg Titer Significantly Predicts Subsequent Relapse Risk in Multiple Sclerosis,” Multiple Sclerosis Journal, 18, 799-806.
Publisher – Google Scholar
Svejgaard, A. (2008). “The Immunogenetics of Multiple Sclerosis,” Immunogenetics, 60, 275-86.
Publisher – Google Scholar
Tejada-Simon, M. V., Zang, Y. C. Q., Hong, J., Rivera, V. M. & Zhang, J. Z. (2003). “Cross-Reactivity with Myelin Basic Protein and Human Herpesvirus-6 in Multiple Sclerosis,” Annals of Neurology, 53, 189-97.
Publisher – Google Scholar
Terasaki, P. I., Park, M. S., Opelz, G. & Ting, A. (1976). “Multiple Sclerosis and High Incidence of A B Lymphocyte Antigen,” Science, 193, 1245-7.
Publisher – Google Scholar
Virtanen, J. O., Wohler, J., Fenton, K., Reich, D. S. & Jacobson, S. (2014). “Oligoclonal Bands in Multiple Sclerosis Reactive Against Two Herpesviruses and Association with Magnetic Resonance Imaging Findings,” Multiple Sclerosis Journal, 20, 27-34.
Publisher – Google Scholar
Wucherpfennig, K. W. & Strominger, J. L. (1995). “Molecular Mimicry in T Cell-Mediated Autoimmunity: Viral Peptides Activate Human T Cell Clones Specific for Myelin Basic Protein,” Cell, 80, 695-705.
Publisher – Google Scholar